bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_5
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
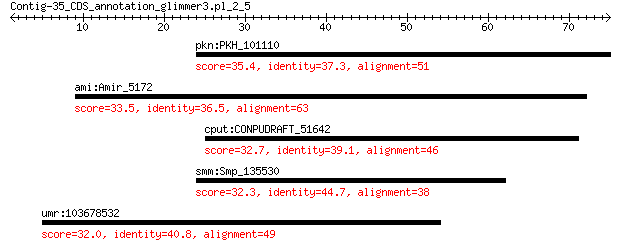
pkn:PKH_101110 hypothetical protein 35.4 0.80
ami:Amir_5172 sensor with HAMP domain 33.5 3.7
cput:CONPUDRAFT_51642 multidrug resistance-associated ABC tran... 32.7 6.5
smm:Smp_135530 leishmanolysin-2 (M08 family) 32.3 7.9
umr:103678532 RNF20; ring finger protein 20, E3 ubiquitin prot... 32.0 9.6
> pkn:PKH_101110 hypothetical protein
Length=1515
Score = 35.4 bits (80), Expect = 0.80, Method: Composition-based stats.
Identities = 19/56 (34%), Positives = 30/56 (54%), Gaps = 5/56 (9%)
Query 24 SMSGLTLLSGRQLTNTPPDKPEEESNPTNL-----KDNNKNTKQKNPYRKDTTFLQ 74
S + + L+SG + N PP P +S+ KDNN+N ++KN + +FLQ
Sbjct 1250 SSTDVNLISGHKSDNMPPKNPPSDSDKIEQGSHSNKDNNQNQEEKNFINEMVSFLQ 1305
> ami:Amir_5172 sensor with HAMP domain
Length=1023
Score = 33.5 bits (75), Expect = 3.7, Method: Composition-based stats.
Identities = 23/66 (35%), Positives = 29/66 (44%), Gaps = 3/66 (5%)
Query 9 TRNGGSAQGKTK--KTRSMSGLTLLSGRQLTNTPPDKPEE-ESNPTNLKDNNKNTKQKNP 65
T+NG + G T S SG +GR L TPP KPE S P + T + P
Sbjct 730 TQNGPTQNGATSGGAPVSESGWAAFAGRALDQTPPAKPEPGRSEPGRSEQGRSETARSEP 789
Query 66 YRKDTT 71
R + T
Sbjct 790 GRSEAT 795
> cput:CONPUDRAFT_51642 multidrug resistance-associated ABC transporter
Length=1693
Score = 32.7 bits (73), Expect = 6.5, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 23/46 (50%), Gaps = 1/46 (2%)
Query 25 MSGLTLLSGRQLTNTPPDKPEEESNPTNLKDNNKNTKQKNPYRKDT 70
+ GL + +G N PDKPEE+S N K N K K +DT
Sbjct 653 IEGLAIENGSFKWNAVPDKPEEDSTSKNAK-NGKKRKHGKDVSRDT 697
> smm:Smp_135530 leishmanolysin-2 (M08 family)
Length=1137
Score = 32.3 bits (72), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 17/38 (45%), Positives = 22/38 (58%), Gaps = 4/38 (11%)
Query 24 SMSGLTLLSGRQLTNTPPDKPEEESNPTNLKDNNKNTK 61
S G+ LL G+Q+TN P+ ES P N DNN N +
Sbjct 760 SSGGIELLVGKQVTNC----PDGESLPINAYDNNINVR 793
> umr:103678532 RNF20; ring finger protein 20, E3 ubiquitin protein
ligase
Length=975
Score = 32.0 bits (71), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 5 VLLYTRNGGSAQGKTKKTRSMSGLTLLSGRQLTNTPPDKPEE-ESNPTNL 53
VL Y R AQ KTR SG LL + T P D+P E + +P +L
Sbjct 494 VLRYKRKLREAQSDLNKTRLRSGTALLQSQSSTEDPKDEPAELKQDPEDL 543
Lambda K H a alpha
0.305 0.124 0.344 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125073504444