bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
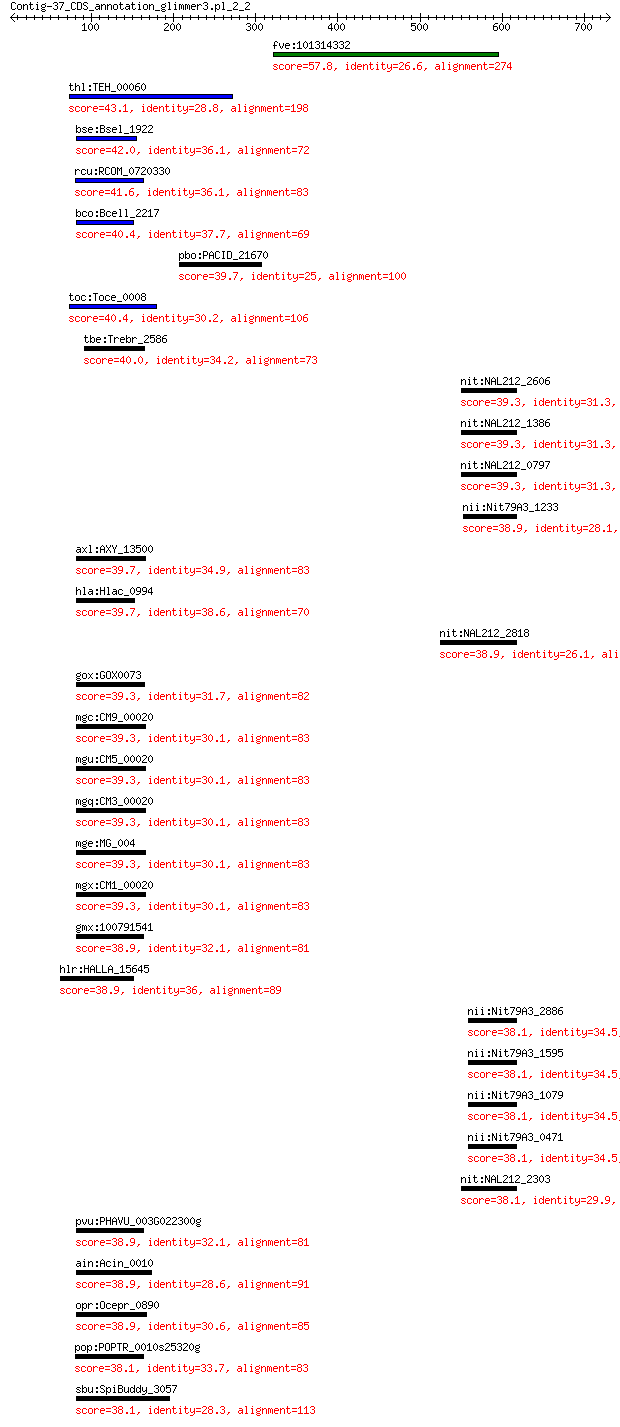
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=731
Score E
Sequences producing significant alignments: (Bits) Value
fve:101314332 capsid protein VP1-like 57.8 6e-06
thl:TEH_00060 gyrA; DNA gyrase subunit A (EC:5.99.1.3) 43.1 0.25
bse:Bsel_1922 DNA topoisomerase IV subunit A (EC:5.99.1.3) 42.0 0.50
rcu:RCOM_0720330 DNA gyrase subunit A, chloroplast/mitochondri... 41.6 0.65
bco:Bcell_2217 DNA topoisomerase IV subunit alpha (EC:5.99.1.3) 40.4 1.5
pbo:PACID_21670 dapB; dihydrodipicolinate reductase (EC:1.3.1.26) 39.7 1.5
toc:Toce_0008 DNA gyrase subunit A (EC:5.99.1.3) 40.4 1.8
tbe:Trebr_2586 DNA gyrase subunit A (EC:5.99.1.3) 40.0 2.2
nit:NAL212_2606 methane monooxygenase/ammonia monooxygenase su... 39.3 2.5
nit:NAL212_1386 methane monooxygenase/ammonia monooxygenase su... 39.3 2.5
nit:NAL212_0797 methane monooxygenase/ammonia monooxygenase su... 39.3 2.5
nii:Nit79A3_1233 methane monooxygenase/ammonia monooxygenase s... 38.9 2.6
axl:AXY_13500 parC; DNA topoisomerase IV subunit A (EC:5.99.1.-) 39.7 2.9
hla:Hlac_0994 DNA gyrase subunit alpha (EC:5.99.1.3) 39.7 3.0
nit:NAL212_2818 methane monooxygenase/ammonia monooxygenase su... 38.9 3.3
gox:GOX0073 DNA topoisomerase IV subunit A (EC:5.99.1.-) 39.3 3.8
mgc:CM9_00020 DNA gyrase subunit A 39.3 3.9
mgu:CM5_00020 DNA gyrase subunit A 39.3 3.9
mgq:CM3_00020 DNA gyrase subunit A 39.3 3.9
mge:MG_004 gyrA; DNA gyrase subunit A (EC:5.99.1.3) 39.3 3.9
mgx:CM1_00020 DNA gyrase subunit A 39.3 4.1
gmx:100791541 DNA gyrase subunit A, chloroplastic/mitochondria... 38.9 4.8
hlr:HALLA_15645 DNA gyrase subunit A 38.9 4.9
nii:Nit79A3_2886 methane monooxygenase/ammonia monooxygenase s... 38.1 4.9
nii:Nit79A3_1595 methane monooxygenase/ammonia monooxygenase s... 38.1 4.9
nii:Nit79A3_1079 methane monooxygenase/ammonia monooxygenase s... 38.1 4.9
nii:Nit79A3_0471 methane monooxygenase/ammonia monooxygenase s... 38.1 4.9
nit:NAL212_2303 methane monooxygenase/ammonia monooxygenase su... 38.1 5.0
pvu:PHAVU_003G022300g hypothetical protein 38.9 5.0
ain:Acin_0010 type IIA topoisomerase 38.9 5.2
opr:Ocepr_0890 DNA gyrase subunit a 38.9 5.5
pop:POPTR_0010s25320g POPTRDRAFT_805680; DNA gyrase subunit A ... 38.1 8.1
sbu:SpiBuddy_3057 DNA gyrase subunit alpha (EC:5.99.1.3) 38.1 8.3
> fve:101314332 capsid protein VP1-like
Length=421
Score = 57.8 bits (138), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 73/289 (25%), Positives = 114/289 (39%), Gaps = 59/289 (20%)
Query 321 AYRFRAYESVYNAYYRDIR-NNPFIVDGRPVYNKWLPSMKGGADNTT----YSLHQCNWE 375
A RAY +YN ++RD N +VD KG +TT Y+L +
Sbjct 177 ALPVRAYNLIYNQWFRDENLQNSVVVD------------KGDGPDTTPSTNYTLLRRGKR 224
Query 376 RDFLTTAVPNPQQGANAPLVGLTVGDVVTRA-------DDGTYSIQKQTVLVDEDGSKYG 428
D+ T+A+P PQ+G A V L +G A D G S + ++ + + G
Sbjct 225 HDYFTSALPWPQKGGTA--VSLPLGTSAPIAFSGASGSDVGVISTTQGNLIKNMYSTGSG 282
Query 429 ISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTIETLRYVNAYQKF 488
S K+ + A YA+L+A T TI LR QK
Sbjct 283 TSLKIG-----------------SATVATGLYADLSAATAA-----TINQLRQSFQIQKL 320
Query 489 LELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSS--- 545
LE + R G Y +I++ + + L PE++GG S +++ + QT + +
Sbjct 321 LERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTP 380
Query 546 QGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ 594
QG A G Y + + E ++IGL++V Y Q
Sbjct 381 QGNLAA--------FGTYMAKGHGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> thl:TEH_00060 gyrA; DNA gyrase subunit A (EC:5.99.1.3)
Length=821
Score = 43.1 bits (100), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 57/214 (27%), Positives = 88/214 (41%), Gaps = 38/214 (18%)
Query 73 ARLNFFKV-TLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----V 127
AR++ V LR + +D DFI N+ D +EP +LP F +L G+ G +G +
Sbjct 125 ARMSRLSVEMLRDLNKDTVDFIGNYDDTEKEPEVLPA--RFPNLLVNGTTGIAVGMATNI 182
Query 128 PTRNAGAEVSLMAHPSQCAANSMLVFSPSVTLQEVCSSFLSNTIPKANCALSFGNSDGRK 187
P N G V AA L+ +P VT E+ P L G S R+
Sbjct 183 PPHNLGEVV---------AAIDRLIENPEVTTNELMEVLPGPDFPTG--GLVMGKSGIRR 231
Query 188 --QVAYFSITLQGYNNAMRSYKLTVGIKKANDKDR---------NSYSGAIVFSSATNVY 236
+ SIT++G K+ + + AN K+R + + I S +
Sbjct 232 AYETGKGSITVRG--------KVEIS-EMANGKERILVTELPYMVNKAKLIERISELHRD 282
Query 237 DRFENVTFVWDESTSSYVTTISFSDKQYSAVSVL 270
R E +T + DES+ + + + SA VL
Sbjct 283 KRIEGITDLRDESSREGMRIVVDVRRDVSASVVL 316
> bse:Bsel_1922 DNA topoisomerase IV subunit A (EC:5.99.1.3)
Length=815
Score = 42.0 bits (97), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 26/80 (33%), Positives = 41/80 (51%), Gaps = 11/80 (14%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAG---- 133
L+ + ++ DF+ NF D EEP +LPGS + +L GS G G +P + G
Sbjct 134 LKDIEKNTVDFMPNFDDSQEEPVVLPGS--YPNLLVNGSTGISSGYATDIPPHHLGEVID 191
Query 134 AEVSLMAHPSQCAANSMLVF 153
A + M HP C+ + +L +
Sbjct 192 AAIHQMDHPD-CSVHDLLTY 210
> rcu:RCOM_0720330 DNA gyrase subunit A, chloroplast/mitochondrial
precursor, putative (EC:5.99.1.3)
Length=540
Score = 41.6 bits (96), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 42/87 (48%), Gaps = 15/87 (17%)
Query 80 VTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAE 135
V L + D DF+ NF D +EP +LP T+L GS G +G +P N G
Sbjct 235 VLLADLELDTVDFVPNFDDSQKEPSLLPA--RLPTLLLNGSSGIAVGMATNIPPHNLGEL 292
Query 136 VSLMAHPSQCAANSMLVFSPSVTLQEV 162
V ++ CA L+ +P TLQE+
Sbjct 293 VDVL-----CA----LIHNPEATLQEL 310
> bco:Bcell_2217 DNA topoisomerase IV subunit alpha (EC:5.99.1.3)
Length=834
Score = 40.4 bits (93), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 26/77 (34%), Positives = 36/77 (47%), Gaps = 10/77 (13%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAG---- 133
L+ + +D DF+ NF D LEEP +LP F +L GS G G +P + G
Sbjct 134 LKDIEKDTVDFMLNFDDSLEEPVVLPS--RFPNLLVNGSTGISSGYATDIPPHHLGEVID 191
Query 134 AEVSLMAHPSQCAANSM 150
A + M HP + M
Sbjct 192 AAIHHMEHPDSTVDDLM 208
> pbo:PACID_21670 dapB; dihydrodipicolinate reductase (EC:1.3.1.26)
Length=246
Score = 39.7 bits (91), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/101 (25%), Positives = 43/101 (43%), Gaps = 4/101 (4%)
Query 207 KLTVGIKKANDKDRNSYSGAIV-FSSATNVYDRFENVTFVWDESTSSYVTTISFSDKQYS 265
+L G+ D+ + IV F+ V D N+++ D + V T F+D++Y
Sbjct 28 ELVAGVDAGEDRSPLEDAEVIVDFTRPDTVMD---NLSWAIDHGIHTVVGTTGFTDERYE 84
Query 266 AVSVLLQTEPGIASSSLWQFGLNTASAVAFVEESETTYETT 306
+ LL PG FG+ + F +E+ YE+
Sbjct 85 ELRALLAGHPGTGCLVAPNFGIGAVLMMHFAQEAARFYESA 125
> toc:Toce_0008 DNA gyrase subunit A (EC:5.99.1.3)
Length=809
Score = 40.4 bits (93), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 32/107 (30%), Positives = 49/107 (46%), Gaps = 8/107 (7%)
Query 73 ARLNFFKVTLRS-MWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRN 131
ARL+ + + S + +D DFI NF D L+EP +LP F +L GS G +G+ T
Sbjct 125 ARLSRIALEMLSDIDKDTVDFIPNFDDTLKEPAVLP--SRFPNLLVNGSSGIAVGMATNI 182
Query 132 AGAEVSLMAHPSQCAANSMLVFSPSVTLQEVCSSFLSNTIPKANCAL 178
+S M++ +P VT QE+ ++ P L
Sbjct 183 PPHNLS-----EVIDGVVMMIENPDVTSQELMAAIKGPDFPTGGIIL 224
> tbe:Trebr_2586 DNA gyrase subunit A (EC:5.99.1.3)
Length=817
Score = 40.0 bits (92), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 11/75 (15%)
Query 91 DFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAHPSQ--CAAN 148
DF+ NF D +EP +LP F +L GS G +G+ T ++ H + C+A
Sbjct 148 DFLPNFDDTRQEPTVLPA--KFPFLLANGSSGIAVGMAT-------NMPPHNLREICSAI 198
Query 149 SMLVFSPSVTLQEVC 163
S + +P V+L+E+C
Sbjct 199 SAYIDNPDVSLEELC 213
> nit:NAL212_2606 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 550 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 609
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 610 QPEFDRI 616
+PEFD++
Sbjct 58 EPEFDKV 64
> nit:NAL212_1386 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 550 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 609
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 610 QPEFDRI 616
+PEFD++
Sbjct 58 EPEFDKV 64
> nit:NAL212_0797 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 550 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 609
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 610 QPEFDRI 616
+PEFD++
Sbjct 58 EPEFDKV 64
> nii:Nit79A3_1233 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.9 bits (89), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 35/64 (55%), Gaps = 0/64 (0%)
Query 553 LGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 612
+ + G G S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTGASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 613 FDRI 616
FD++
Sbjct 61 FDKV 64
> axl:AXY_13500 parC; DNA topoisomerase IV subunit A (EC:5.99.1.-)
Length=816
Score = 39.7 bits (91), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 40/87 (46%), Gaps = 15/87 (17%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
LR + +D D+I NF D LEEP ILP F +L GS G G +P N +
Sbjct 134 LRDINKDTVDYIPNFDDSLEEPVILPA--RFPNLLVNGSTGISAGYATDIPPHNLTEVID 191
Query 138 LMAHPSQCAANSMLVFSPSVTLQEVCS 164
A L+ P VT++E+ +
Sbjct 192 ---------AVIKLIDKPDVTVEELMN 209
> hla:Hlac_0994 DNA gyrase subunit alpha (EC:5.99.1.3)
Length=827
Score = 39.7 bits (91), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 27/78 (35%), Positives = 38/78 (49%), Gaps = 11/78 (14%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAG---- 133
L + D DF SN+ D LEEP +LP + F +L GS G +G +P N G
Sbjct 145 LADIERDTVDFTSNYDDRLEEPEVLPAA--FPNLLVNGSSGIAVGMSTNIPPHNLGEVID 202
Query 134 AEVSLMAHPSQCAANSML 151
A V+L+ P C ++
Sbjct 203 ATVALIKEPD-CTVEDLM 219
> nit:NAL212_2818 methane monooxygenase/ammonia monooxygenase
subunit C
Length=295
Score = 38.9 bits (89), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 12/92 (13%)
Query 525 ISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLL 584
I+ L + ++Q D +T+ G+ SG A S + ++ ++ D + Y +G+L
Sbjct 11 INNYLRIFLIKQMEDMMATTY--------GTTSGAA----SANYDMSLWYDSKYYKLGML 58
Query 585 TVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRI 616
T+ V I+ + F Y+ +D +PEFD++
Sbjct 59 TMLLVAIFWIWYQRTFAYSHGMDSMEPEFDKV 90
> gox:GOX0073 DNA topoisomerase IV subunit A (EC:5.99.1.-)
Length=736
Score = 39.3 bits (90), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 43/86 (50%), Gaps = 15/86 (17%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
L + +D DF + + ++ EP +LPG+ F +L G+ G +G +P NA AE+
Sbjct 136 LDGIQDDAVDFRATYDNEDHEPIVLPGT--FPNLLANGAAGIAVGMATSIPPHNA-AEI- 191
Query 138 LMAHPSQCAANSMLVFSPSVTLQEVC 163
C A +L+ P VT+ E+
Sbjct 192 -------CRAAKLLIEKPDVTIAELV 210
> mgc:CM9_00020 DNA gyrase subunit A
Length=836
Score = 39.3 bits (90), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (52%), Gaps = 7/83 (8%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + +D DFI+N+ + +EP +LP + F +L GS G +G+ T +S +
Sbjct 147 LKDIDKDTVDFIANYDGEEKEPTVLPAA--FPNLLANGSSGIAVGMSTSIPSHNLSEL-- 202
Query 142 PSQCAANSMLVFSPSVTLQEVCS 164
A ML+ +P T QE+ +
Sbjct 203 ---IAGLIMLIDNPQCTFQELLT 222
> mgu:CM5_00020 DNA gyrase subunit A
Length=836
Score = 39.3 bits (90), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (52%), Gaps = 7/83 (8%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + +D DFI+N+ + +EP +LP + F +L GS G +G+ T +S +
Sbjct 147 LKDIDKDTVDFIANYDGEEKEPTVLPAA--FPNLLANGSSGIAVGMSTSIPSHNLSEL-- 202
Query 142 PSQCAANSMLVFSPSVTLQEVCS 164
A ML+ +P T QE+ +
Sbjct 203 ---IAGLIMLIDNPQCTFQELLT 222
> mgq:CM3_00020 DNA gyrase subunit A
Length=836
Score = 39.3 bits (90), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (52%), Gaps = 7/83 (8%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + +D DFI+N+ + +EP +LP + F +L GS G +G+ T +S +
Sbjct 147 LKDIDKDTVDFIANYDGEEKEPTVLPAA--FPNLLANGSSGIAVGMSTSIPSHNLSEL-- 202
Query 142 PSQCAANSMLVFSPSVTLQEVCS 164
A ML+ +P T QE+ +
Sbjct 203 ---IAGLIMLIDNPQCTFQELLT 222
> mge:MG_004 gyrA; DNA gyrase subunit A (EC:5.99.1.3)
Length=836
Score = 39.3 bits (90), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (52%), Gaps = 7/83 (8%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + +D DFI+N+ + +EP +LP + F +L GS G +G+ T +S +
Sbjct 147 LKDIDKDTVDFIANYDGEEKEPTVLPAA--FPNLLANGSSGIAVGMSTSIPSHNLSEL-- 202
Query 142 PSQCAANSMLVFSPSVTLQEVCS 164
A ML+ +P T QE+ +
Sbjct 203 ---IAGLIMLIDNPQCTFQELLT 222
> mgx:CM1_00020 DNA gyrase subunit A
Length=836
Score = 39.3 bits (90), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (52%), Gaps = 7/83 (8%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + +D DFI+N+ + +EP +LP + F +L GS G +G+ T +S +
Sbjct 147 LKDIDKDTVDFIANYDGEEKEPTVLPAA--FPNLLANGSSGIAVGMSTSIPSHNLSEL-- 202
Query 142 PSQCAANSMLVFSPSVTLQEVCS 164
A ML+ +P T QE+ +
Sbjct 203 ---IAGLIMLIDNPQCTFQELLT 222
> gmx:100791541 DNA gyrase subunit A, chloroplastic/mitochondrial-like
Length=935
Score = 38.9 bits (89), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 41/85 (48%), Gaps = 15/85 (18%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
L + +D DF+ NF + +EP +LP T+L GS G +G +P N G V
Sbjct 219 LTDLEQDTVDFVPNFDNSQKEPSLLPA--RLPTLLLNGSSGIAVGMATNIPPHNLGEVVD 276
Query 138 LMAHPSQCAANSMLVFSPSVTLQEV 162
++ +L+ +P TLQE+
Sbjct 277 VLC---------VLIHNPEATLQEL 292
> hlr:HALLA_15645 DNA gyrase subunit A
Length=833
Score = 38.9 bits (89), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 45/99 (45%), Gaps = 15/99 (15%)
Query 62 PMVFP--VQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTG 119
P P + RM A + L + +D DF SN+ D LEEP +LP S F +L G
Sbjct 124 PAAAPRYTEARMSA---LSEELLEDIDKDTVDFQSNYDDRLEEPTVLPSS--FPNLLVNG 178
Query 120 SLGDYLG----VPTRNAG----AEVSLMAHPSQCAANSM 150
S G +G +P N G A + L+ +P + M
Sbjct 179 SSGIAVGMSTNIPPHNLGEVIDATIELIDNPDSTVEDLM 217
> nii:Nit79A3_2886 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 559 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 612
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 613 FDRI 616
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_1595 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 559 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 612
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 613 FDRI 616
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_1079 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 559 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 612
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 613 FDRI 616
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_0471 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 559 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 612
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 613 FDRI 616
FD++
Sbjct 61 FDKV 64
> nit:NAL212_2303 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 20/67 (30%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 550 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 609
A G+ SG A S + ++ ++ D + Y +G+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKLGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 610 QPEFDRI 616
+PEFD++
Sbjct 58 EPEFDKV 64
> pvu:PHAVU_003G022300g hypothetical protein
Length=942
Score = 38.9 bits (89), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 41/85 (48%), Gaps = 15/85 (18%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
L + +D DF+ NF + +EP +LP T+L GS G +G +P N G V
Sbjct 226 LADLEQDTVDFVPNFDNSQKEPSLLPA--RLPTLLLNGSSGIAVGMATNIPPHNLGEVVD 283
Query 138 LMAHPSQCAANSMLVFSPSVTLQEV 162
++ +L+ +P TLQE+
Sbjct 284 VLC---------VLIHNPEATLQEL 299
> ain:Acin_0010 type IIA topoisomerase
Length=842
Score = 38.9 bits (89), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 42/95 (44%), Gaps = 15/95 (16%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
LR + +D DFI N+ + L+EP +LP +L GS G +G +P N G V
Sbjct 147 LRDIEKDTVDFIPNYDESLKEPTVLP--SKVPALLINGSAGIAVGMATNIPPHNLGEVVD 204
Query 138 LMAHPSQCAANSMLVFSPSVTLQEVCSSFLSNTIP 172
+ ML+ P T++++ + P
Sbjct 205 ALV---------MLIDHPDATIEDLMGAIKGPDFP 230
> opr:Ocepr_0890 DNA gyrase subunit a
Length=804
Score = 38.9 bits (89), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 39/89 (44%), Gaps = 15/89 (17%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAEVS 137
LR + +D DF+ NF EEP +LP F +L GS G +G +P N G ++
Sbjct 131 LRDIDKDTVDFVENFDGTAEEPTVLPA--GFPNLLANGSTGIAVGMATSIPPHNLGELIN 188
Query 138 LMAHPSQCAANSMLVFSPSVTLQEVCSSF 166
+A ++ P +TL V
Sbjct 189 ALA---------AMIDDPEITLDGVMQHL 208
> pop:POPTR_0010s25320g POPTRDRAFT_805680; DNA gyrase subunit
A family protein
Length=948
Score = 38.1 bits (87), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 42/87 (48%), Gaps = 15/87 (17%)
Query 80 VTLRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLG----VPTRNAGAE 135
V L + +D DF+ NF + +EP + P T+L GS G +G +P N G
Sbjct 225 VFLADLEQDTVDFVPNFDNSQKEPSLFP--TRLPTLLLNGSSGIAVGMATKIPPHNLGEL 282
Query 136 VSLMAHPSQCAANSMLVFSPSVTLQEV 162
V ++ CA L+ +P TLQE+
Sbjct 283 VDVL-----CA----LIHNPEATLQEL 300
> sbu:SpiBuddy_3057 DNA gyrase subunit alpha (EC:5.99.1.3)
Length=855
Score = 38.1 bits (87), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 55/121 (45%), Gaps = 18/121 (15%)
Query 82 LRSMWEDYSDFISNFRDDLEEPYILPGSQNFTTMLGTGSLGDYLGVPTRNAGAEVSLMAH 141
L+ + ++ +F N+ D ++EP +LP S F +L GS G +G+ T ++ H
Sbjct 136 LQDIQKETVNFGPNYDDSMQEPTVLPAS--FPFLLANGSSGIAVGMAT-------NMAPH 186
Query 142 PSQ--CAANSMLVFSPSVTLQEVCSSFLSNTIPKAN--CALSFGNSD----GRKQVAYFS 193
Q C A S ++ +P +T+ E+ P C L G D GR ++ S
Sbjct 187 NLQEICDAISAVIDNPDITIDELMEHIKGPDFPSGGIICGLQ-GIKDAFMTGRGKIVVRS 245
Query 194 I 194
+
Sbjct 246 V 246
Lambda K H a alpha
0.318 0.133 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1768716362272