bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_7
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
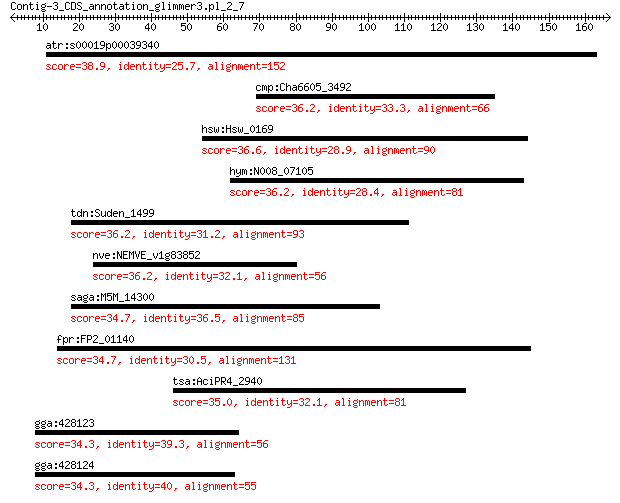
atr:s00019p00039340 AMTR_s00019p00039340; hypothetical protein 38.9 0.25
cmp:Cha6605_3492 hypothetical protein 36.2 0.58
hsw:Hsw_0169 peptidase u62 modulator of DNA gyrase 36.6 2.1
hym:N008_07105 hypothetical protein 36.2 2.3
tdn:Suden_1499 resistance-nodulation-cell division family tran... 36.2 2.6
nve:NEMVE_v1g83852 hypothetical protein 36.2 2.7
saga:M5M_14300 hypothetical protein 34.7 6.6
fpr:FP2_01140 Predicted transcriptional regulator with C-termi... 34.7 6.6
AciPR4_2940 conjugative relaxase domain-containing protein 35.0 7.1
gga:428123 MADPRT, ART7.2; ADP-ribosyltransferase 1 (EC:2.4.2.31) 34.3 8.9
gga:428124 ART7B; GPI-anchored ADP-ribosyltransferase 34.3 9.8
> atr:s00019p00039340 AMTR_s00019p00039340; hypothetical protein
Length=259
Score = 38.9 bits (89), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 64/153 (42%), Gaps = 6/153 (4%)
Query 11 FGCGFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHA 70
CG R P + +PST +G V +EQ R + V + D +L +A
Sbjct 76 LACGLRLLDP---LFVYPSTILGREKAIVVNLEQIR--CIITADEVLFLAFVDNYVLRYA 130
Query 71 ADLAKRAGVSTVQRFLDSKSPRSSSLQEQL-DKLNPSDDELLSMVKSRHLQHPSEILAWI 129
L KR + DS +P L L K P DEL S + + HL+ + + +
Sbjct 131 QLLQKRLTMKNDMGVGDSWAPVGGDLAAALVIKAYPLSDELTSKISTLHLEGVRRLKSRL 190
Query 130 DSINELAEDMRSEALKQTAENGSICPIVLVKKK 162
++ + +R E + ++G + + L KKK
Sbjct 191 VALTRRVQKVRDEIEQLMDDDGDMAEMYLTKKK 223
> cmp:Cha6605_3492 hypothetical protein
Length=95
Score = 36.2 bits (82), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 22/67 (33%), Positives = 36/67 (54%), Gaps = 1/67 (1%)
Query 69 HAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQHPSEI-LA 127
H DLA + TV++ ++ S ++Q+ D LNPS E+ + +S L PSE+ L
Sbjct 12 HILDLAIKHNDKTVKQLVNYPSSLLIAMQQYKDNLNPSYTEIYKIFESGLLLSPSEVDLN 71
Query 128 WIDSINE 134
W+ + N
Sbjct 72 WLKNQNN 78
> hsw:Hsw_0169 peptidase u62 modulator of DNA gyrase
Length=441
Score = 36.6 bits (83), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 26/90 (29%), Positives = 40/90 (44%), Gaps = 3/90 (3%)
Query 54 ESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSM 113
SVS +D L+ A KRAGV+T F D+ R E++ +L P D E + +
Sbjct 42 NSVSTAGATDNVSLVVEARFGKRAGVATCNEFDDATLRRCVQRAEEIARLAPEDPEYMPL 101
Query 114 VKSRHLQHPSEILAWIDSINELAEDMRSEA 143
+ + P A +I D R++A
Sbjct 102 LGPQQYLTPVSYAAGTAAIT---PDFRAQA 128
> hym:N008_07105 hypothetical protein
Length=441
Score = 36.2 bits (82), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 41/81 (51%), Gaps = 4/81 (5%)
Query 62 SDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQH 121
S+VS+ + A KR+GV+T +F D+ R E++ +L P D E + M+ ++
Sbjct 51 SNVSLAVEA-RFGKRSGVATCNQFDDATLRRCVQRAEEIARLAPEDPEYMPMLGAQQYLT 109
Query 122 PSEILAWIDSINELAEDMRSE 142
P+ A S + D R++
Sbjct 110 PTTYAA---STAGITPDFRAQ 127
> tdn:Suden_1499 resistance-nodulation-cell division family transporter
Length=1005
Score = 36.2 bits (82), Expect = 2.6, Method: Composition-based stats.
Identities = 29/98 (30%), Positives = 48/98 (49%), Gaps = 6/98 (6%)
Query 18 FTPTREISTFPSTRIGEISLE---VDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLA 74
F ++ +TF + EI E D ++ F + FGE S +L + V+ ++ D
Sbjct 728 FLEQKQSTTFNERGVMEIKTEDIKKDSIDTFLNFSIPFGEGKSVKL-TQVADIIEIRDYE 786
Query 75 K--RAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDEL 110
K + S ++ F + R ++ QE LDKL P+ DEL
Sbjct 787 KINKLNGSIIKTFFATIDKRKTTSQEVLDKLEPTLDEL 824
> nve:NEMVE_v1g83852 hypothetical protein
Length=796
Score = 36.2 bits (82), Expect = 2.7, Method: Composition-based stats.
Identities = 18/57 (32%), Positives = 31/57 (54%), Gaps = 1/57 (2%)
Query 24 ISTFPSTRIGEISLEVDPVEQFRFETETFGESV-SYRLRSDVSMLLHAADLAKRAGV 79
+S P + L+ DPVE F T+TF ++ SY +++ SM LH ++ + G+
Sbjct 627 VSVLPGKAVQPADLDYDPVEDAIFWTDTFTGTINSYNMQTRKSMTLHRCNVERPDGI 683
> saga:M5M_14300 hypothetical protein
Length=244
Score = 34.7 bits (78), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 31/87 (36%), Positives = 43/87 (49%), Gaps = 9/87 (10%)
Query 18 FTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRA 77
F I+ +R+ E+ L + V ET Y L S S A AKRA
Sbjct 90 FPAASGINNHMGSRLTEMVLPMRAV------METLAPRRLYFLDSKTSRRSVAWQEAKRA 143
Query 78 GVSTVQR--FLDSKSPRSSSLQEQLDK 102
G+ TVQR FLD++ P ++++Q QLDK
Sbjct 144 GLETVQRDVFLDNE-PSAAAIQVQLDK 169
> fpr:FP2_01140 Predicted transcriptional regulator with C-terminal
CBS domains
Length=184
Score = 34.7 bits (78), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 63/141 (45%), Gaps = 17/141 (12%)
Query 14 GFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADL 73
GFR T ++ ++ +LE P ++ E+ S + R V +L+ AD
Sbjct 39 GFRVLTQSK-------IKVIADALETTPSYIMGWDEESHQNEWSSKFRDSVMQILNNADP 91
Query 74 A--KRAGVST--VQRFLDSKSPRSSSLQEQL-DKLNPSDDELLS-----MVKSRHLQHPS 123
A K AG+S ++ LD P S + + D+L S D LL M+K+ Q
Sbjct 92 ADLKAAGISVQEIEEELDGSEPISLAAACSIADQLGESLDSLLGHTPKEMIKAALQQEDG 151
Query 124 EILAWIDSINELAEDMRSEAL 144
+ ID + +L+ D R EAL
Sbjct 152 QTAEIIDLLLDLSADRRQEAL 172
> tsa:AciPR4_2940 conjugative relaxase domain-containing protein
Length=907
Score = 35.0 bits (79), Expect = 7.1, Method: Composition-based stats.
Identities = 26/90 (29%), Positives = 42/90 (47%), Gaps = 14/90 (16%)
Query 46 RFETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTV----QRFLDSKSPRSSSLQEQLD 101
+F T + ++YRLR+ L A A R+G V Q +LD+ SPRS ++E L+
Sbjct 207 QFATAVYQSELTYRLRN-----LGYAIEAGRSGAPEVKGYSQEYLDASSPRSQQIREHLE 261
Query 102 KLNPSDDELLSMV-----KSRHLQHPSEIL 126
K E + + + P+E+L
Sbjct 262 KSGYQGPEAAQIAAHSTRDRKQIHSPAEVL 291
> gga:428123 MADPRT, ART7.2; ADP-ribosyltransferase 1 (EC:2.4.2.31)
Length=296
Score = 34.3 bits (77), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 22/57 (39%), Positives = 29/57 (51%), Gaps = 11/57 (19%)
Query 8 ELCFGCGFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSY-RLRSD 63
E C+G P +E STFP GE + + P EQFR T+ E S+ +LRS
Sbjct 202 ETCYG------VPIKEFSTFP----GEDEVLIPPFEQFRVTNSTYTEGRSFIQLRSQ 248
> gga:428124 ART7B; GPI-anchored ADP-ribosyltransferase
Length=302
Score = 34.3 bits (77), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 29/56 (52%), Gaps = 11/56 (20%)
Query 8 ELCFGCGFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSY-RLRS 62
E C+G P +E STFP GE + + P EQFR T+ E S+ +LRS
Sbjct 204 ETCYG------VPIKEFSTFP----GEDEVLIPPFEQFRVTNSTYTEGRSFIQLRS 249
Lambda K H a alpha
0.315 0.130 0.362 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125369324234