bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-41_CDS_annotation_glimmer3.pl_2_2
Length=582
Score E
Sequences producing significant alignments: (Bits) Value
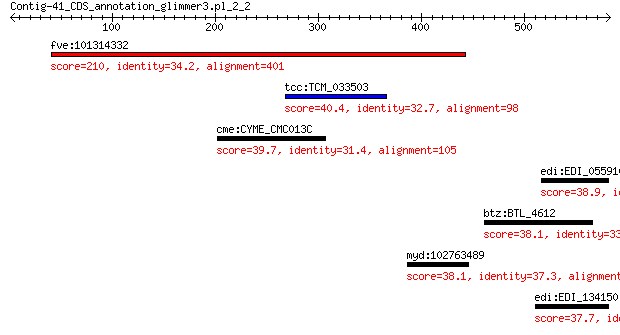
fve:101314332 capsid protein VP1-like 210 2e-58
tcc:TCM_033503 Proton pump interactor 1-like protein 40.4 0.87
cme:CYME_CMC013C 26S proteasome regulatory subunit RPN11 39.7 1.4
edi:EDI_055910 hypothetical protein 38.9 3.1
btz:BTL_4612 short chain dehydrogenase family protein 38.1 4.8
myd:102763489 STOX1; storkhead box 1 38.1
edi:EDI_134150 hypothetical protein 37.7 8.4
> fve:101314332 capsid protein VP1-like
Length=421
Score = 210 bits (535), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 137/409 (33%), Positives = 210/409 (51%), Gaps = 72/409 (18%)
Query 41 VLPGDSFNISTSKVVRSQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREFCGENREGAWAP 100
VLPGD+FN++ + R T + P+MDN+ LD+++FFVPNRLVW +W +F GE + A
Sbjct 77 VLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGE--QDNPAD 134
Query 101 TVEYTVPSIAPPPGGFAKGTIADYMGLPIGVEWKATDPLAPSALPFRGYALICNEFFRDE 160
++ Y++P P GG+A G++ DY GLP + ++ ++ SALP R Y LI N++FRDE
Sbjct 135 SISYSIPQQVSPAGGYAVGSLQDYFGLPTAGQVGVSNTVSHSALPVRAYNLIYNQWFRDE 194
Query 161 NLTDPLLISLDDANQQGSNGDDYANDVANGGKPFKAARMHDYFSSCLPSSQK-GSSVGIP 219
NL + +++ GD + + + HDYF+S LP QK G++V +P
Sbjct 195 NLQNSVVV---------DKGDGPDTTPSTNYTLLRRGKRHDYFTSALPWPQKGGTAVSLP 245
Query 220 IHVPGFAGGTFPVTTTDNWTVPASAVPAVFGLFTTPLNGTIDGRTAYPASTGSSELEQGQ 279
+ G + P+ + A + G+ +T I + Y +G+S L+ G
Sbjct 246 L------GTSAPIAFS-------GASGSDVGVISTTQGNLI--KNMYSTGSGTS-LKIGS 289
Query 280 KIFVSDGHSAPGPMCWMAGSNITYARNVWSPINLSTTIPAAGSGDDVDASFTVNELRLAF 339
T A +++ ++ +T + T+N+LR +F
Sbjct 290 A---------------------TVATGLYADLSAAT-------------AATINQLRQSF 315
Query 340 AYQRFLESMARNGSRYTELLLGLFGVRSPDARLQRPEYLGGNRIPISVSEV-------TN 392
Q+ LE AR G+RYTE++ FGV SPDARLQRPEYLGG PI+++ +
Sbjct 316 QIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQ 375
Query 393 NAQTPEDYLGDLGAKSSTGDVNHDFIKSFTEHGYLFGLFVVRYDHSYSQ 441
TP+ L G + G H F +SF EHG++ GL VR D +Y Q
Sbjct 376 GTTTPQGNLAAFGTYMAKG---HGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> tcc:TCM_033503 Proton pump interactor 1-like protein
Length=374
Score = 40.4 bits (93), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 49/102 (48%), Gaps = 15/102 (15%)
Query 268 ASTGSSELEQGQKIFVSDG-HSAPGPMCWMA-GSNITYARNVWSPINLSTTIPAAGSGDD 325
A S EL+Q +KI + D P W+ GSN N+ + LST IP D
Sbjct 72 ARKQSEELDQQKKILIDDKRQEMVSPRVWVTHGSN-----NLANEEQLSTEIPNVQQKD- 125
Query 326 VDASFTVNEL--RLAFAYQRFLESMARNGSRYTELLLGLFGV 365
+++SF E+ R F Y SM+RN ++ ++L G+ +
Sbjct 126 INSSFLGREINNRAYFWY-----SMSRNKEQFVQILEGVMQI 162
> cme:CYME_CMC013C 26S proteasome regulatory subunit RPN11
Length=325
Score = 39.7 bits (91), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 33/111 (30%), Positives = 47/111 (42%), Gaps = 16/111 (14%)
Query 202 YFSS-CLPSSQKGSSVGIPIHVPGFAGGTFPVTTTDNWTVPASAVPAVFGLFTTPLNGTI 260
Y SS L K + G+P+ V G G F D+WT+ V F P +GT
Sbjct 33 YLSSLALLKILKHARAGVPMEVMGLLLGEF----VDDWTIN------VVDYFAMPQSGTG 82
Query 261 DGRTAYPASTGSSELEQGQK-----IFVSDGHSAPGPMCWMAGSNITYARN 306
A A LE Q+ + GHS PG CW++G ++ A++
Sbjct 83 VSVEAIDAVYQQQFLEALQQTGRHEVVCGWGHSHPGFGCWLSGVDVNTAQS 133
> edi:EDI_055910 hypothetical protein
Length=546
Score = 38.9 bits (89), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 5/65 (8%)
Query 516 AHWHLADHYAKVPTLSDGWIREDKANVDRVLAVQSSVADQFWCDLWISNMCTRPMPMYSI 575
HW + DHY K+PT WI+E ++ L + + +C ++ N+ T MP +
Sbjct 55 VHWMITDHYRKLPT---EWIKEYYEPLEHYLKSHPEHSKEVYC--YMMNITTDIMPSIIL 109
Query 576 PGSLD 580
P +D
Sbjct 110 PSKID 114
> btz:BTL_4612 short chain dehydrogenase family protein
Length=328
Score = 38.1 bits (87), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 35/130 (27%), Positives = 55/130 (42%), Gaps = 26/130 (20%)
Query 461 AHLGETGVYQAEIMATPENMADPTKVFGFQEIWADYRYKPSLVTG--------------- 505
AH T +A I ++AD T V F + AD + ++
Sbjct 55 AHAIRTRYPRARIEVEALDLADLTSVCRFADAVADRHGRVDILCNNAGVMFLPLRRTRDG 114
Query 506 -EMRPGVTNSLAHWHL---------ADHYAKVPTLSDGWIREDKANVDRVLAVQSSVADQ 555
EM+ G TN L H+ L A H A+V T+S G+ R K +D +LA + +
Sbjct 115 FEMQMG-TNHLGHFALTGLLLPALRASHRARVVTMSSGFNRLGKIRLDNMLAERGYNKYR 173
Query 556 FWCDLWISNM 565
+CD ++N+
Sbjct 174 AYCDSKLANL 183
> myd:102763489 STOX1; storkhead box 1
Length=881
Score = 38.1 bits (87), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 22/69 (32%), Positives = 36/69 (52%), Gaps = 10/69 (14%)
Query 386 SVSEVTNNAQTPEDYLGDLGAKS---STGDVNHDFIKSFTEHGYLFGLFV-------VRY 435
S + ++ A+TP+D+LGD G K+ S N D K FT+ LF +++
Sbjct 751 SSRKASSEAETPQDFLGDTGKKAACWSQSAQNQDMRKRFTQKLELFNTTHMPEMAQDIQH 810
Query 436 DHSYSQGIE 444
+HS+ +G E
Sbjct 811 EHSHLEGTE 819
> edi:EDI_134150 hypothetical protein
Length=546
Score = 37.7 bits (86), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 21/71 (30%), Positives = 35/71 (49%), Gaps = 6/71 (8%)
Query 510 GVTNSLAHWHLADHYAKVPTLSDGWIREDKANVDRVLAVQSSVADQFWCDLWISNMCTRP 569
VT S+ HW + DHY +PT WI+E ++ L + + +C ++ N+ T
Sbjct 50 SVTKSV-HWMITDHYHNLPT---EWIKEYYEPLEHYLKSHPEHSKEVYC--YMMNITTDV 103
Query 570 MPMYSIPGSLD 580
MP +P +D
Sbjct 104 MPSVILPSKID 114
Lambda K H a alpha
0.319 0.136 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1335954814206