bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-43_CDS_annotation_glimmer3.pl_2_1
Length=306
Score E
Sequences producing significant alignments: (Bits) Value
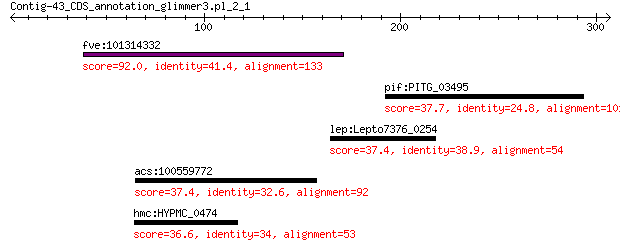
fve:101314332 capsid protein VP1-like 92.0 3e-18
pif:PITG_03495 hypothetical protein 37.7 3.2
lep:Lepto7376_0254 urea ABC transporter ATP-binding protein (E... 37.4 3.3
acs:100559772 hmgcs1; 3-hydroxy-3-methylglutaryl-CoA synthase ... 37.4 4.1
hmc:HYPMC_0474 acxB; acetone carboxylase subunit alpha (EC:6.4... 36.6 7.0
> fve:101314332 capsid protein VP1-like
Length=421
Score = 92.0 bits (227), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 71/134 (53%), Gaps = 1/134 (1%)
Query 38 GGVSTWVPMEARLDNATSATINQLRQAISVQQYYEALARGGSRYREQIRAIWDVIISDKT 97
G + + A L AT+ATINQLRQ+ +Q+ E ARGG+RY E IR+ + V D
Sbjct 288 GSATVATGLYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDAR 347
Query 98 VQVPEYLGGGRYHVNINQI-VQTSGQQTSNDTPIGETGAMSVTPINESSFTKSFEEHGFV 156
+Q PEYLGGG +NI I TP G A F++SF EHG V
Sbjct 348 LQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHV 407
Query 157 IGVCCVRHNRSYQQ 170
IG+ VR + +YQQ
Sbjct 408 IGLVSVRADLTYQQ 421
> pif:PITG_03495 hypothetical protein
Length=1020
Score = 37.7 bits (86), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 25/105 (24%), Positives = 52/105 (50%), Gaps = 8/105 (8%)
Query 192 LGEQPVKKKEIMLTGDTTDDETFGYQEAWADYRMKPNRVSGLMRSNATGTLEFWHYADNY 251
L +P++++ GDT+D + AD R+ P R++ ++R+N L H + +Y
Sbjct 340 LDYEPLERELSWKEGDTSDRQIVVTSFPRADPRLPPRRLALVLRNNKGAPLNGAHASTSY 399
Query 252 AKVPTLSQEWMAE----GKEEIARTLIVEDEPQFFGAIRIANKTT 292
++ +L+ ++ E +E + L + D F I +A++ T
Sbjct 400 VEITSLTDVYLGEVNFATREPLESVLRIPD----FSFIELASRDT 440
> lep:Lepto7376_0254 urea ABC transporter ATP-binding protein
(EC:3.6.3.28)
Length=374
Score = 37.4 bits (85), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 21/57 (37%), Positives = 30/57 (53%), Gaps = 3/57 (5%)
Query 164 HNRSYQQGLERFWSRKDRLDYYVPQFANLGEQPVKKK---EIMLTGDTTDDETFGYQ 217
H S Q+ L+ + +RK LD VP+ NLGE+ K EI+ + + DE YQ
Sbjct 292 HRFSLQKELQNYLARKIPLDELVPKLQNLGEKEKVLKLSYEIIRSNEINKDELMVYQ 348
> acs:100559772 hmgcs1; 3-hydroxy-3-methylglutaryl-CoA synthase
1 (soluble)
Length=564
Score = 37.4 bits (85), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 44/93 (47%), Gaps = 8/93 (9%)
Query 65 ISVQQYYEALARGGSRYREQIRAIWDVIISDKTVQVPEYLGGGRYHVNINQIVQTS-GQQ 123
+S+Q Y AL R S YR++IR W +DK + ++ G +H ++VQ S +
Sbjct 287 LSIQCYLSALDRCYSVYRKKIRTQWQKEGTDKDFNLNDF-GFMIFHSPYCKLVQKSVARL 345
Query 124 TSNDTPIGETGAMSVTPINESSFTKSFEEHGFV 156
ND I + TPINE+ E G V
Sbjct 346 LLNDFLIDQ------TPINENGIYNGLEAFGDV 372
> hmc:HYPMC_0474 acxB; acetone carboxylase subunit alpha (EC:6.4.1.6)
Length=775
Score = 36.6 bits (83), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/57 (32%), Positives = 34/57 (60%), Gaps = 6/57 (11%)
Query 64 AISVQQYYEALARGGSRYREQIRAIWDV----IISDKTVQVPEYLGGGRYHVNINQI 116
A VQQ + A + G R+ ++ RA WD+ IS++ + +P Y G RY ++++++
Sbjct 710 ATQVQQMFAASFKLGPRFEKEFRAFWDLPESWTISEEELGIPSY--GSRYSMDLSEL 764
Lambda K H a alpha
0.315 0.132 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 524805453876