bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-48_CDS_annotation_glimmer3.pl_2_1
Length=473
Score E
Sequences producing significant alignments: (Bits) Value
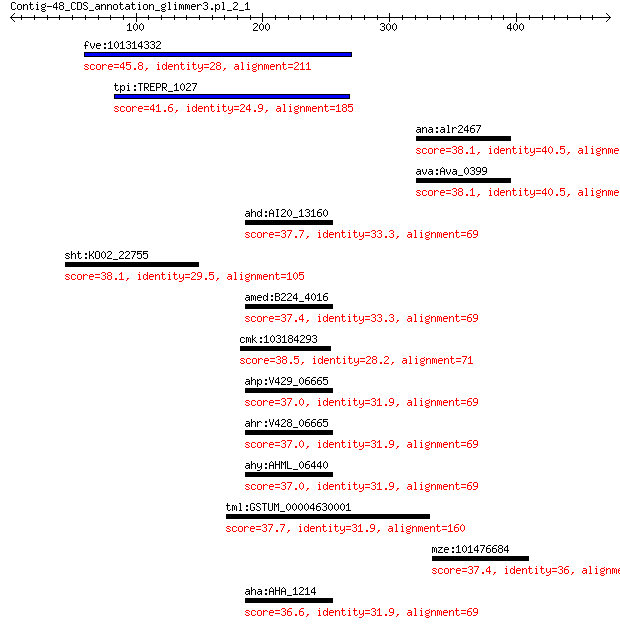
fve:101314332 capsid protein VP1-like 45.8 0.015
tpi:TREPR_1027 hypothetical protein 41.6 0.49
ana:alr2467 hypothetical protein 38.1 2.6
ava:Ava_0399 hypothetical protein 38.1 2.8
ahd:AI20_13160 glutathione S-transferase 37.7 3.1
sht:KO02_22755 hypothetical protein 38.1 3.6
amed:B224_4016 glutathione S-transferase, N-terminal 37.4 4.0
cmk:103184293 clip1; CAP-GLY domain containing linker protein 1 38.5
ahp:V429_06665 glutathione S-transferase 37.0 5.1
ahr:V428_06665 glutathione S-transferase 37.0 5.1
ahy:AHML_06440 glutathione S-transferase 37.0 5.1
tml:GSTUM_00004630001 hypothetical protein 37.7 5.9
mze:101476684 NADPH oxidase 4-like 37.4 6.7
aha:AHA_1214 glutathione S-transferase 36.6 7.1
> fve:101314332 capsid protein VP1-like
Length=421
Score = 45.8 bits (107), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 59/211 (28%), Positives = 83/211 (39%), Gaps = 22/211 (10%)
Query 59 FPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLK 118
P+ AY IY +FR + +S VD G PD T T + L
Sbjct 178 LPVRAYNLIYNQWFRDENLQ----NSVVVDKGDG---------PDTTPS-----TNYTLL 219
Query 119 YCNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSS 178
D LP Q G T +G +D IS T L S
Sbjct 220 RRGKRHDYFTSALPWPQKGGTAVSLPLGTSAPIAFSGASGSDVGVIS-TTQGNLIKNMYS 278
Query 179 SGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQI 238
+G T +TVA + L +DLSA + ++ LRQ+ +Q+ E G + Y E I
Sbjct 279 TGSGTSLKIGSATVA--TGLYADLSAATAA-TINQLRQSFQIQKLLERDARGGTRYTEII 335
Query 239 RKHFGVNLPQSLSNLCTYIGGISRNLDISEV 269
R HFGV P + Y+GG S ++I+ +
Sbjct 336 RSHFGVASPDARLQRPEYLGGGSTPINIAPI 366
> tpi:TREPR_1027 hypothetical protein
Length=1039
Score = 41.6 bits (96), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 46/206 (22%), Positives = 87/206 (42%), Gaps = 27/206 (13%)
Query 83 SSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYCNWNKDM---LMGILPDSQFGEC 139
++ D Y VS L T ++ +D ++ FD + W + L G+ +S+ G
Sbjct 478 TALKADAYDRVSDKLKTFEDEFAADLFRRRDEFDTRLGKWKDSLDSELTGLAEESREGRR 537
Query 140 CYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSSGLSTPGVTSGSTV-ALKSPL 198
+ FE +L+ F D + +S + + G+ + T+ LK L
Sbjct 538 KLEQNFE---DELQRRFADEDARMVSELEHLKAEAGAFEEGIRNQLAQADETLGTLKDQL 594
Query 199 ISDL--------SALQSQFSVLALRQAEALQRW-KEISQ-----SGDSDYREQIRKHFGV 244
+L S+++++ +L AE L+++ +EI + S ++R FG
Sbjct 595 DRNLDDARLSAESSVKAELGRFSLSMAETLKQYQREIEEDLRKISAQVEFRS---GEFGE 651
Query 245 NLPQSLSNLCTYIG---GISRNLDIS 267
L +S N+ + G G R+LD+S
Sbjct 652 LLDESRRNIDEWQGKFAGQMRDLDVS 677
> ana:alr2467 hypothetical protein
Length=263
Score = 38.1 bits (87), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 41/87 (47%), Gaps = 15/87 (17%)
Query 321 GQDGQLLVTDAESLP--IPEFDNIGMEVLP-----------MTQIFNSPKASIVNLFNAG 367
G D LLVT A +P + F GM V+P + ++ N+PKA+I+NL
Sbjct 179 GIDQVLLVTSALHMPRSLKIFQRQGMNVIPAPTDFLVSEGELQELGNTPKAAILNLLPDT 238
Query 368 YNPRYFNWKTKLDVVNGAFTTTLKSWV 394
YN F K V G+F L+ WV
Sbjct 239 YNLHLFTNALKEYV--GSFVYWLRGWV 263
> ava:Ava_0399 hypothetical protein
Length=263
Score = 38.1 bits (87), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 41/87 (47%), Gaps = 15/87 (17%)
Query 321 GQDGQLLVTDAESLP--IPEFDNIGMEVLP-----------MTQIFNSPKASIVNLFNAG 367
G D LLVT A +P + F GM V+P + ++ N+PKA+I+NL
Sbjct 179 GIDKVLLVTSALHMPRSLKIFQRQGMNVIPAPTDFLVSAGELQELGNTPKAAILNLLPDT 238
Query 368 YNPRYFNWKTKLDVVNGAFTTTLKSWV 394
YN F K V G+F L+ WV
Sbjct 239 YNLHLFTNALKEYV--GSFIYWLRGWV 263
> ahd:AI20_13160 glutathione S-transferase
Length=208
Score = 37.7 bits (86), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (55%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDY--REQIRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ LQR K ++ +G + E +R H
Sbjct 8 VGTGSTWAMRAALVLAMSGLEWQEQVFDLEDAKDLQRLKRLAPAGLVPWLDHEGVRVHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L LC
Sbjct 68 LAIAEYLHELC 78
> sht:KO02_22755 hypothetical protein
Length=305
Score = 38.1 bits (87), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 44/110 (40%), Gaps = 22/110 (20%)
Query 44 TGYTQQYIQNNYVNLFPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPD 103
+G T Y +N +NL + Q+ + F+W SGVSS +TVLP
Sbjct 212 SGKTHCY-KNEKINLKAFVRLQQAPSNLFKWQ---------------SGVSS-FITVLPS 254
Query 104 YTSDYWKSDTMFDL-----KYCNWNKDMLMGILPDSQFGECCYKSIFETP 148
T+ W D + K + L+ LPD + CY IF P
Sbjct 255 CTAIRWNKDLFATMNSTLEKVVSQVPVGLLQCLPDEAAAQLCYNRIFNKP 304
> amed:B224_4016 glutathione S-transferase, N-terminal
Length=208
Score = 37.4 bits (85), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYRE--QIRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ALQR K + +G + + ++R H
Sbjct 8 VGTGSTWAMRASLVLAMSGLEWQEQVFDLEDAKALQRLKRTAPAGLVPWLDDGELRIHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L LC
Sbjct 68 LAIAEYLHELC 78
> cmk:103184293 clip1; CAP-GLY domain containing linker protein
1
Length=2200
Score = 38.5 bits (88), Expect = 4.4, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 182 STPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIRKH 241
ST G S +AL SPL+ ++S+L++Q + + L WK + DS +++ +++
Sbjct 519 STKGSGSTDDLALPSPLLEEVSSLRNQITAMNNEHLSQLNAWKYKLAASDSHHQQDLQQL 578
Query 242 FGVNLPQSLSN 252
N S+ N
Sbjct 579 RATNDTLSMDN 589
> ahp:V429_06665 glutathione S-transferase
Length=208
Score = 37.0 bits (84), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQ--IRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ LQR K ++ +G + + +R H
Sbjct 8 VGTGSTWAMRAALVLAMSGLEWQEQVFDLEDAKELQRLKRLAPAGLVPWLDHDGVRVHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L +LC
Sbjct 68 LAIAEYLHDLC 78
> ahr:V428_06665 glutathione S-transferase
Length=208
Score = 37.0 bits (84), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQ--IRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ LQR K ++ +G + + +R H
Sbjct 8 VGTGSTWAMRAALVLAMSGLEWQEQVFDLEDAKELQRLKRLAPAGLVPWLDHDGVRVHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L +LC
Sbjct 68 LAIAEYLHDLC 78
> ahy:AHML_06440 glutathione S-transferase
Length=208
Score = 37.0 bits (84), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQ--IRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ LQR K ++ +G + + +R H
Sbjct 8 VGTGSTWAMRAALVLAMSGLEWQEQVFDLEDAKELQRLKRLAPAGLVPWLDHDGVRVHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L +LC
Sbjct 68 LAIAEYLHDLC 78
> tml:GSTUM_00004630001 hypothetical protein
Length=450
Score = 37.7 bits (86), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 51/176 (29%), Positives = 72/176 (41%), Gaps = 37/176 (21%)
Query 171 PLTTENSSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSG 230
P+T S+ S PG GST +P L+ Q F +R EA Q WK G
Sbjct 27 PITPLGSTPYSSRPGSLPGSTSGFSAP---GLNVPQRYFHSRRVRPGEAQQPWK-----G 78
Query 231 DSDYREQ---IRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLA-------AEGDT 280
D +E+ I G+ + +++ + G +S VVN+ + G
Sbjct 79 KKDPKEKWVTIIPCIGLAVGLAIAGFFVWHG-------VSSVVNHKYCPVLIEDWSNGFD 131
Query 281 AVIAGKGV---GAGNGSFTYTT--DEHCVVMCIY-HAVPLLDYTITGQDGQLLVTD 330
I K V G GNG F YTT +E+ V + H P L QD +L+ TD
Sbjct 132 EKIWTKEVECGGFGNGQFDYTTNSEENSYVKGGHLHIRPTL------QDEKLITTD 181
> mze:101476684 NADPH oxidase 4-like
Length=559
Score = 37.4 bits (85), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 32/75 (43%), Gaps = 8/75 (11%)
Query 334 LPIPEFDNIGMEVLPMTQIFNSPKASIVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSW 393
LP P D ME+LPM Q PK + F G + FN+ L V G T
Sbjct 367 LPEPRID---MEILPMAQKRRYPKVYVDGPF-GGPSEEVFNYDVSLCVAGGIGVTPFAC- 421
Query 394 VSPVTESLLSGWFGF 408
V +LL GW GF
Sbjct 422 ---VLRALLDGWMGF 433
> aha:AHA_1214 glutathione S-transferase
Length=208
Score = 36.6 bits (83), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 39/71 (55%), Gaps = 2/71 (3%)
Query 186 VTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQ--IRKHFG 243
V +GST A+++ L+ +S L+ Q V L A+ LQR K ++ +G + + +R H
Sbjct 8 VGTGSTWAMRAALVLAMSGLEWQEQVFDLEDAKELQRLKRLAPAGLVPWLDHDGVRVHDS 67
Query 244 VNLPQSLSNLC 254
+ + + L LC
Sbjct 68 LAIAEYLHELC 78
Lambda K H a alpha
0.317 0.134 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1018243303440