bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_2
Length=598
Score E
Sequences producing significant alignments: (Bits) Value
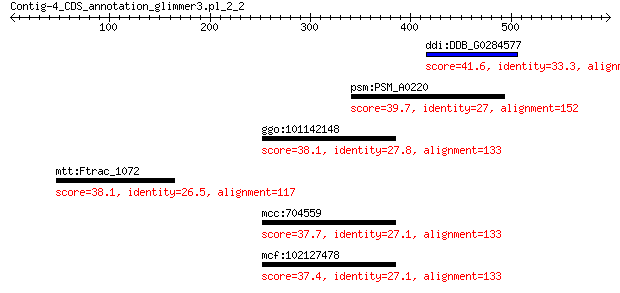
ddi:DDB_G0284577 NOT2/NOT3/NOT5 family protein 41.6 0.59
psm:PSM_A0220 sdaC; serine transport protein 39.7 1.7
ggo:101142148 AMPH; amphiphysin isoform 1 38.1
mtt:Ftrac_1072 xanthan lyase xalb 38.1 7.9
mcc:704559 AMPH; amphiphysin 37.7 8.2
mcf:102127478 AMPH; amphiphysin 37.4 9.8
> ddi:DDB_G0284577 NOT2/NOT3/NOT5 family protein
Length=579
Score = 41.6 bits (96), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 30/92 (33%), Positives = 47/92 (51%), Gaps = 11/92 (12%)
Query 416 VGSSQGSEKFEARDWGVLMCIYHNVPLLDYVSSAPDPQFFVTQNTDLPIPELDSIGMQSV 475
+G QG E+++ +D+ ++ LL +S V +NTDL I L SIG+
Sbjct 343 LGYDQGHEEYQRKDFNIVDKDNGKYGLLGLLS--------VIKNTDLDISTL-SIGIDLT 393
Query 476 PLAMYTNSDGELVSGFVSP--DYTMGYLPRYF 505
L + NS GE+ + F SP DY++ P Y+
Sbjct 394 ALGLNLNSQGEISNSFGSPWVDYSISRKPEYY 425
> psm:PSM_A0220 sdaC; serine transport protein
Length=423
Score = 39.7 bits (91), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 41/165 (25%), Positives = 73/165 (44%), Gaps = 20/165 (12%)
Query 341 RRGEALQRWKEISLNVPQNYRAQIKAHFGVDVGENMS---------GMSTYVGGDSSSLD 391
RG L R+ S N ++ ++ HFGV+ G +S + Y G ++++D
Sbjct 74 HRG--LARFVLSSKNKEADFTDVVEEHFGVNAGRLISLLYFLSIFPILLIYGVGLTNTVD 131
Query 392 ISEVVNTNLQS-GDVASEAVIAGKGVGSSQGSEKFEARDWGVLMCIYHNVPLLDYVSSAP 450
V ++S VA ++ +G G E+ R + +L +Y V +L ++S
Sbjct 132 SFMVNQLGMESPSRVALSGILVALMIGLMMGGERLMLRAFAIL--VYPLVGILFFLSLYL 189
Query 451 DPQFFVTQNTDLPIPELDSIGMQ---SVPLAMYTNSDGELVSGFV 492
P + Q D +PEL S S+P+ +++ S +S FV
Sbjct 190 IPSW---QMPDTALPELGSFSKTLWLSIPIIVFSFSHAAAISSFV 231
> ggo:101142148 AMPH; amphiphysin isoform 1
Length=695
Score = 38.1 bits (87), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 59/137 (43%), Gaps = 14/137 (10%)
Query 252 DMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWASGSPSSKAPVVVGAAASSPNFTIRA 311
++ +G P GD A G TS++PE + + K P+ AP + A+ + R
Sbjct 558 EITIGAEPKETTGDAAPPGPTSETPELATEQKPIQDPQPTPSAPAM---GAADQLASARE 614
Query 312 ESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVD 371
S + P + V+T A + D L L+RG ++ L VP + A A + V
Sbjct 615 ASQELPPGFLYKVETLHDFEAANSDELTLQRG-------DVVLVVPSDSEADQDAGWLVG 667
Query 372 VGE----NMSGMSTYVG 384
V E ++TY G
Sbjct 668 VKESDWLQYRDLATYKG 684
> mtt:Ftrac_1072 xanthan lyase xalb
Length=1110
Score = 38.1 bits (87), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 53/124 (43%), Gaps = 8/124 (6%)
Query 47 FTLKRQHFTRTQPVNTSAYTRIREYYDWFWVPLHLLWRHAPEVISQMQSNVQ------HA 100
+ KR F T P+ +AYT++ E + + P + W I Q+ +Q A
Sbjct 556 YPFKRLQFVET-PLQFAAYTKVYETHQAYLQPETVFWPEKGGDIRQLDLRMQLRDMNNQA 614
Query 101 GSQTSSLTLGNYLPTISSSQLSAVCSRLFGKKNYF-GYDRSDLSYKLMQYLRVGNSGQVS 159
SQ +LT + + + V ++ G F GY++ D +Y + N+G VS
Sbjct 615 KSQNQTLTDKQKQANVFNDLIKRVITKQIGANYTFDGYNQDDANYSIFPNYYTYNTGFVS 674
Query 160 VNFG 163
+G
Sbjct 675 EEWG 678
> mcc:704559 AMPH; amphiphysin
Length=652
Score = 37.7 bits (86), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 60/137 (44%), Gaps = 14/137 (10%)
Query 252 DMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWASGSPSSKAPVVVGAAASSPNFTIRA 311
++ +G P+ D A +G TS++PE + + K P AP + A + ++R
Sbjct 515 EITIGAEPEETTEDTAPLGPTSETPELATEPKPLQDPQPMPSAPALGTADQLA---SVRE 571
Query 312 ESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVD 371
S + P + V+T A + D L L+RG ++ L VP + A A + V
Sbjct 572 ASQELPPGFLYKVETLHDFEAANSDELTLQRG-------DVVLVVPSDSEADQDAGWLVG 624
Query 372 VGE----NMSGMSTYVG 384
V E ++TY G
Sbjct 625 VKESDWLQYRDLATYKG 641
> mcf:102127478 AMPH; amphiphysin
Length=694
Score = 37.4 bits (85), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 60/137 (44%), Gaps = 14/137 (10%)
Query 252 DMFMGVFPDTQFGDVATIGITSDSPESSLQLKAWASGSPSSKAPVVVGAAASSPNFTIRA 311
++ +G P+ D A +G TS++PE + + K P AP + A + ++R
Sbjct 557 EITIGAEPEETTEDTAPLGPTSETPELATEPKPLQDPQPMPSAPALGTADQLA---SVRE 613
Query 312 ESGNMNPANILGVDTSSLSLAGSFDVLALRRGEALQRWKEISLNVPQNYRAQIKAHFGVD 371
S + P + V+T A + D L L+RG ++ L VP + A A + V
Sbjct 614 ASQELPPGFLYKVETLHDFEAANSDELTLQRG-------DVVLVVPSDSEADQDAGWLVG 666
Query 372 VGE----NMSGMSTYVG 384
V E ++TY G
Sbjct 667 VKESDWLQYRDLATYKG 683
Lambda K H a alpha
0.318 0.132 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1375374696596