bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_6
Length=68
Score E
Sequences producing significant alignments: (Bits) Value
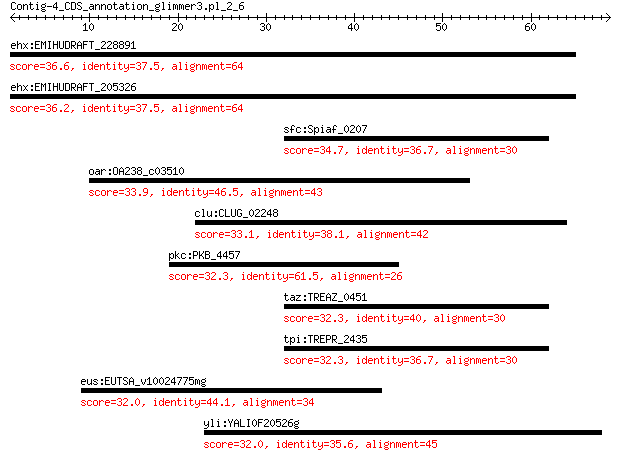
ehx:EMIHUDRAFT_228891 hypothetical protein 36.6 0.36
ehx:EMIHUDRAFT_205326 hypothetical protein 36.2 0.37
sfc:Spiaf_0207 family 3 adenylate cyclase 34.7 1.4
oar:OA238_c03510 Asp/Glu/hydantoin racemase-like protein 33.9 1.6
clu:CLUG_02248 hypothetical protein 33.1 3.8
pkc:PKB_4457 transcriptional regulator, LysR family 32.3 5.9
taz:TREAZ_0451 adenylate/guanylate cyclase catalytic domain-co... 32.3 6.2
tpi:TREPR_2435 adenylate/guanylate cyclase catalytic domain-co... 32.3 6.3
eus:EUTSA_v10024775mg hypothetical protein 32.0 8.1
yli:YALI0F20526g YALI0F20526p 32.0 8.4
> ehx:EMIHUDRAFT_228891 hypothetical protein
Length=1200
Score = 36.6 bits (83), Expect = 0.36, Method: Composition-based stats.
Identities = 24/68 (35%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 1 MEKKLRFVNVVACSNLGTRIRLTLSSE---TDDLRQLVADALIRIGQYGSSQETF-LVQH 56
+E +L V S LG R+R ++ D+L Q VAD L+ G YG++ T ++
Sbjct 227 LETRLALKAAVLASPLGGRLRKANRADKLDADELAQQVADGLLFAGGYGTTHLTLAALER 286
Query 57 IDSAPPLY 64
I S P LY
Sbjct 287 ISSDPALY 294
> ehx:EMIHUDRAFT_205326 hypothetical protein
Length=1219
Score = 36.2 bits (82), Expect = 0.37, Method: Composition-based stats.
Identities = 24/68 (35%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 1 MEKKLRFVNVVACSNLGTRIRLTLSSE---TDDLRQLVADALIRIGQYGSSQETF-LVQH 56
+E +L V S LG R+R ++ D+L Q VAD L+ G YG++ T ++
Sbjct 242 LETRLALKAAVLASPLGGRLRKANRADKLDADELAQQVADGLLFAGGYGTTHLTLAALER 301
Query 57 IDSAPPLY 64
I S P LY
Sbjct 302 ISSDPALY 309
> sfc:Spiaf_0207 family 3 adenylate cyclase
Length=910
Score = 34.7 bits (78), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/30 (37%), Positives = 23/30 (77%), Gaps = 0/30 (0%)
Query 32 RQLVADALIRIGQYGSSQETFLVQHIDSAP 61
R ++AD L+R+G++G++ F +++ID +P
Sbjct 76 RSILADGLLRLGEFGAAAAIFDIEYIDPSP 105
> oar:OA238_c03510 Asp/Glu/hydantoin racemase-like protein
Length=248
Score = 33.9 bits (76), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 20/47 (43%), Positives = 30/47 (64%), Gaps = 4/47 (9%)
Query 10 VVACSNLG-TRIRLT---LSSETDDLRQLVADALIRIGQYGSSQETF 52
+ AC++LG TRI L ++S +D LR ++ADA IR+ + S E F
Sbjct 116 IAACAHLGVTRIGLISPYVASVSDQLRVVLADAGIRVTHFASFDEPF 162
> clu:CLUG_02248 hypothetical protein
Length=1121
Score = 33.1 bits (74), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/46 (35%), Positives = 26/46 (57%), Gaps = 4/46 (9%)
Query 22 LTLSSET----DDLRQLVADALIRIGQYGSSQETFLVQHIDSAPPL 63
LTL+S D + Q DAL++ GQ+ +++ + H+DSA P
Sbjct 538 LTLASRLNIVDDTVYQKAVDALMKFGQFEKAEDLLFMAHVDSAVPF 583
> pkc:PKB_4457 transcriptional regulator, LysR family
Length=299
Score = 32.3 bits (72), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/26 (62%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 19 RIRLTLSSETDDLRQLVADALIRIGQ 44
R+RL S E DLRQ +ADA +RIGQ
Sbjct 123 RLRLLASLEVIDLRQGLADAAVRIGQ 148
> taz:TREAZ_0451 adenylate/guanylate cyclase catalytic domain-containing
protein
Length=905
Score = 32.3 bits (72), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 32 RQLVADALIRIGQYGSSQETFLVQHIDSAP 61
R ++AD L+R+ +YG F +++ID+AP
Sbjct 71 RSVMADGLLRLKEYGVETAIFDIEYIDNAP 100
> tpi:TREPR_2435 adenylate/guanylate cyclase catalytic domain-containing
protein
Length=879
Score = 32.3 bits (72), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 11/30 (37%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 32 RQLVADALIRIGQYGSSQETFLVQHIDSAP 61
R ++AD L+R+ +YG+ F +++ID +P
Sbjct 33 RSIMADGLLRLKEYGADAAIFDIEYIDKSP 62
> eus:EUTSA_v10024775mg hypothetical protein
Length=578
Score = 32.0 bits (71), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 22/34 (65%), Gaps = 0/34 (0%)
Query 9 NVVACSNLGTRIRLTLSSETDDLRQLVADALIRI 42
NV A S L T I + L SE +DL+ +V+D+ R+
Sbjct 497 NVAANSQLNTEISVGLKSEPNDLKGIVSDSSNRV 530
> yli:YALI0F20526g YALI0F20526p
Length=1939
Score = 32.0 bits (71), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 16/45 (36%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 23 TLSSETDDLRQLVADALIRIGQYGSSQETFLVQHIDSAPPLYDMR 67
TLSS+T +L+ + + + Y S+ ET LV+H ++A L +R
Sbjct 1049 TLSSQTAELKNQLQETAVNCATYQSNYETELVKHAEAANSLTALR 1093
Lambda K H a alpha
0.321 0.135 0.373 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127465902564