bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_8
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
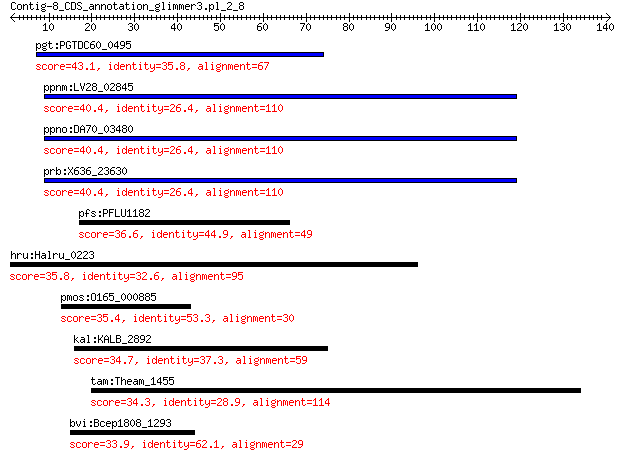
pgt:PGTDC60_0495 hypothetical protein 43.1 0.001
ppnm:LV28_02845 ATPase AAA 40.4 0.053
ppno:DA70_03480 ATPase AAA 40.4 0.053
prb:X636_23630 ATPase AAA 40.4 0.054
pfs:PFLU1182 hypothetical protein 36.6 0.99
hru:Halru_0223 hypothetical protein 35.8 1.7
pmos:O165_000885 hypothetical protein 35.4 3.3
kal:KALB_2892 hypothetical protein 34.7 6.2
tam:Theam_1455 hypothetical protein 34.3 7.3
bvi:Bcep1808_1293 TP901 family phage tail tape measure protein 33.9 9.8
> pgt:PGTDC60_0495 hypothetical protein
Length=103
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/69 (35%), Positives = 39/69 (57%), Gaps = 5/69 (7%)
Query 7 GAAAGAAAGSVVPGIGTAIGAIGGAAISAIGNWFGNRSNRKASREA--FERESKFAREER 64
GAA GAA GSV+PG+GT G I GAA+ ++ G ++N A F ++++F + +
Sbjct 37 GAAIGAAIGSVIPGLGTITGGIAGAAVESL---IGEKANENIDNFADNFFQDTEFEFQCK 93
Query 65 LAQQQWIEQ 73
+W +
Sbjct 94 SCSHKWTRK 102
> ppnm:LV28_02845 ATPase AAA
Length=541
Score = 40.4 bits (93), Expect = 0.053, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 47/111 (42%), Gaps = 2/111 (2%)
Query 9 AAGAAAGSVVPGIGTAI-GAIGGAAISAIGNWFGNRSNRKASREAFERESKFAREERLAQ 67
A A V+P + + G + A I W R A R ++ E+ A
Sbjct 74 AYNAPQARVIPSLDKLVPGLVAYLARDVIDGWLYQRHRDGALLPWLVRRLRYVEPEQGAP 133
Query 68 QQWIEQMYEKNNSYNSPAAQMQRLKDAGLNPDLMYSRGDVGNATAPEAPAQ 118
I+ + S NS + QRL+ +G+ L+++R D+ N T PE A+
Sbjct 134 YVVIDMLANTMQSANSNLTE-QRLRRSGMTTSLVFTRDDIANRTIPELLAE 183
> ppno:DA70_03480 ATPase AAA
Length=541
Score = 40.4 bits (93), Expect = 0.053, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 47/111 (42%), Gaps = 2/111 (2%)
Query 9 AAGAAAGSVVPGIGTAI-GAIGGAAISAIGNWFGNRSNRKASREAFERESKFAREERLAQ 67
A A V+P + + G + A I W R A R ++ E+ A
Sbjct 74 AYNAPQARVIPSLDKLVPGLVAYLARDVIDGWLYQRHRDGALLPWLVRRLRYVEPEQGAP 133
Query 68 QQWIEQMYEKNNSYNSPAAQMQRLKDAGLNPDLMYSRGDVGNATAPEAPAQ 118
I+ + S NS + QRL+ +G+ L+++R D+ N T PE A+
Sbjct 134 YVVIDMLANTMQSANSNLTE-QRLRRSGMTTSLVFTRDDIANRTIPELLAE 183
> prb:X636_23630 ATPase AAA
Length=541
Score = 40.4 bits (93), Expect = 0.054, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 47/111 (42%), Gaps = 2/111 (2%)
Query 9 AAGAAAGSVVPGIGTAI-GAIGGAAISAIGNWFGNRSNRKASREAFERESKFAREERLAQ 67
A A V+P + + G + A I W R A R ++ E+ A
Sbjct 74 AYNAPQARVIPSLDKLVPGLVAYLARDVIDGWLYQRHRDGALLPWLVRRLRYVEPEQGAP 133
Query 68 QQWIEQMYEKNNSYNSPAAQMQRLKDAGLNPDLMYSRGDVGNATAPEAPAQ 118
I+ + S NS + QRL+ +G+ L+++R D+ N T PE A+
Sbjct 134 YVVIDMLANTMQSANSNLTE-QRLRRSGMTTSLVFTRDDIANRTIPELLAE 183
> pfs:PFLU1182 hypothetical protein
Length=383
Score = 36.6 bits (83), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 22/54 (41%), Positives = 31/54 (57%), Gaps = 5/54 (9%)
Query 17 VVPGIGTAI-GAIGGAAISAIGNWFGNR----SNRKASREAFERESKFAREERL 65
V+PG+GTA+ GAIGG S G W G++ S+R S A +E AR + +
Sbjct 275 VLPGVGTAVGGAIGGLLGSEAGAWLGDKLFGSSDRLPSPNAVSKELNHARPDNV 328
> hru:Halru_0223 hypothetical protein
Length=322
Score = 35.8 bits (81), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 46/96 (48%), Gaps = 19/96 (20%)
Query 1 MPSPAAGAAAGAAAGSVVPGIGTAIG-AIGGAAISAIGNWFGNRSNRKASREAFERESKF 59
+ S GAA GAA GS+ PG+GT G A+G AA +G++ ++ S
Sbjct 226 LASMGTGAAVGAAVGSIFPGLGTEAGAAVGTAASGVVGDFV---------KQGIGWASDN 276
Query 60 AREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKDAG 95
+E+ A +EQMYE+ + + LKD G
Sbjct 277 LVDEKAAS---LEQMYEEISRM------YEELKDEG 303
> pmos:O165_000885 hypothetical protein
Length=762
Score = 35.4 bits (80), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/31 (52%), Positives = 21/31 (68%), Gaps = 1/31 (3%)
Query 13 AAGSVVPGIGTAIGA-IGGAAISAIGNWFGN 42
A GSVVPG+GT +G+ +GG A+G W G
Sbjct 456 ALGSVVPGLGTVVGSTVGGMVGEAVGGWVGK 486
> kal:KALB_2892 hypothetical protein
Length=632
Score = 34.7 bits (78), Expect = 6.2, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 34/59 (58%), Gaps = 5/59 (8%)
Query 16 SVVPGIGTAIGAIGGAAISAIGNWFGNRSNRKASREAFERESKFAREERLAQQQWIEQM 74
S++PGIG AIG + G SA+G FG R+ + S EA + + R+ LA + +E +
Sbjct 490 SLLPGIGAAIGGVLG---SALGTLFGARNGYRTSMEASREQQR--RQRALALRHELEPL 543
> tam:Theam_1455 hypothetical protein
Length=668
Score = 34.3 bits (77), Expect = 7.3, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 50/116 (43%), Gaps = 6/116 (5%)
Query 20 GIGTAIGAIGGAAISA--IGNWFGNRSNRKASREAFERESKFAREERLAQQQWIEQMYEK 77
G+ +A G I G + I G+++ A R E K A+E L ++ + K
Sbjct 142 GLPSASGVIRGNSAEQPFIAAKSGSKTGELARRGVVEDGVKNAKETELGKKAVSNRFVNK 201
Query 78 NNSYNSPAAQMQRLKDAGLNPDLMYSRGDVGNATAPEAPAQAPTPRYNVIPTNTYG 133
+ AA + AG+N + GNA AP + P PR N +P N+YG
Sbjct 202 SLLPAPAAAFAPEVLAAGVNVKELLK----GNACAPHGEGELPAPRGNFLPHNSYG 253
> bvi:Bcep1808_1293 TP901 family phage tail tape measure protein
Length=754
Score = 33.9 bits (76), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 18/29 (62%), Positives = 20/29 (69%), Gaps = 0/29 (0%)
Query 15 GSVVPGIGTAIGAIGGAAISAIGNWFGNR 43
GSVVP GTA+GAIGGA G+ FG R
Sbjct 677 GSVVPIAGTAVGAIGGAIAGYFGHDFGER 705
Lambda K H a alpha
0.311 0.126 0.367 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126935320330