bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_5
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
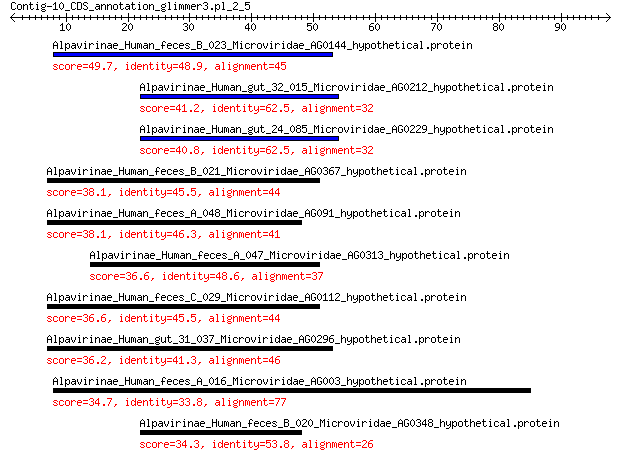
Alpavirinae_Human_feces_B_023_Microviridae_AG0144_hypothetical.... 49.7 6e-11
Alpavirinae_Human_gut_32_015_Microviridae_AG0212_hypothetical.p... 41.2 5e-08
Alpavirinae_Human_gut_24_085_Microviridae_AG0229_hypothetical.p... 40.8 6e-08
Alpavirinae_Human_feces_B_021_Microviridae_AG0367_hypothetical.... 38.1 5e-07
Alpavirinae_Human_feces_A_048_Microviridae_AG091_hypothetical.p... 38.1 6e-07
Alpavirinae_Human_feces_A_047_Microviridae_AG0313_hypothetical.... 36.6 2e-06
Alpavirinae_Human_feces_C_029_Microviridae_AG0112_hypothetical.... 36.6 2e-06
Alpavirinae_Human_gut_31_037_Microviridae_AG0296_hypothetical.p... 36.2 3e-06
Alpavirinae_Human_feces_A_016_Microviridae_AG003_hypothetical.p... 34.7 1e-05
Alpavirinae_Human_feces_B_020_Microviridae_AG0348_hypothetical.... 34.3 2e-05
> Alpavirinae_Human_feces_B_023_Microviridae_AG0144_hypothetical.protein
Length=87
Score = 49.7 bits (117), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 22/45 (49%), Positives = 36/45 (80%), Gaps = 2/45 (4%)
Query 8 VKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIES 52
VKDLF ++R T +EE ++++ IG+HLATE +F T+EEA+ +I++
Sbjct 3 VKDLF-VVRAT-NEENNDFIVTIGRHLATEKKFKTKEEAQAYIDT 45
> Alpavirinae_Human_gut_32_015_Microviridae_AG0212_hypothetical.protein
Length=72
Score = 41.2 bits (95), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 20/32 (63%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 22 EQEEYVIVIGKHLATEARFSTREEAEQFIESK 53
E+E+YVI I LA++ F+TREEAEQ+IESK
Sbjct 15 EEEKYVICIKDELASKEEFNTREEAEQYIESK 46
> Alpavirinae_Human_gut_24_085_Microviridae_AG0229_hypothetical.protein
Length=72
Score = 40.8 bits (94), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 20/32 (63%), Positives = 26/32 (81%), Gaps = 0/32 (0%)
Query 22 EQEEYVIVIGKHLATEARFSTREEAEQFIESK 53
E+E+YVI I LA++ F+TREEAEQ+IESK
Sbjct 15 EEEKYVICIKDELASKEEFNTREEAEQYIESK 46
> Alpavirinae_Human_feces_B_021_Microviridae_AG0367_hypothetical.protein
Length=79
Score = 38.1 bits (87), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 28/44 (64%), Gaps = 3/44 (7%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFI 50
+KD F I+R E +E++I IG HLATE +F +R+ AE I
Sbjct 3 NLKDAF-IVRPLP--ESDEFIITIGNHLATEEKFKSRKAAEMRI 43
> Alpavirinae_Human_feces_A_048_Microviridae_AG091_hypothetical.protein
Length=79
Score = 38.1 bits (87), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 27/41 (66%), Gaps = 3/41 (7%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAE 47
+KD F I+R E +E++I IG HLATE +F +R+ AE
Sbjct 3 NLKDAF-IVRPLP--ESDEFIITIGNHLATEEKFKSRKAAE 40
> Alpavirinae_Human_feces_A_047_Microviridae_AG0313_hypothetical.protein
Length=75
Score = 36.6 bits (83), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 18/37 (49%), Positives = 25/37 (68%), Gaps = 1/37 (3%)
Query 14 IIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFI 50
+IR T +E E ++I IG HLATE +F +R+ AE I
Sbjct 9 VIRPTDTDENE-FIITIGNHLATEEKFKSRKAAEMRI 44
> Alpavirinae_Human_feces_C_029_Microviridae_AG0112_hypothetical.protein
Length=75
Score = 36.6 bits (83), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 28/44 (64%), Gaps = 2/44 (5%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFI 50
+K+ F I T T+E E++I IG HLATE +F +R+ AE I
Sbjct 3 NLKEAFVIRPTDTNEN--EFMITIGNHLATEEKFKSRKAAEMRI 44
> Alpavirinae_Human_gut_31_037_Microviridae_AG0296_hypothetical.protein
Length=79
Score = 36.2 bits (82), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 27/46 (59%), Gaps = 2/46 (4%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIES 52
D F IIR +EE +++I +G HLAT F + +EAE IE+
Sbjct 7 NFNDAF-IIRAL-NEENNDFIITLGDHLATPEHFKSYDEAENSIEA 50
> Alpavirinae_Human_feces_A_016_Microviridae_AG003_hypothetical.protein
Length=76
Score = 34.7 bits (78), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/77 (34%), Positives = 48/77 (62%), Gaps = 2/77 (3%)
Query 8 VKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIESKdydliwaliaalid 67
V++ F+I+ T T E+ +YVIV G A++ F+T + A+++I +K +DLI+ + A
Sbjct 2 VENFFKILPTNT--EENDYVIVCGNVQASKEHFNTVKAAQEYINTKPWDLIFTISYAAHK 59
Query 68 aKDVQEMEAQQEKDVQK 84
A + QE E + + ++K
Sbjct 60 ALNAQEAEENKSETIKK 76
> Alpavirinae_Human_feces_B_020_Microviridae_AG0348_hypothetical.protein
Length=81
Score = 34.3 bits (77), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 14/26 (54%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 22 EQEEYVIVIGKHLATEARFSTREEAE 47
E EEY+I IG HLATE +F + + A+
Sbjct 15 ESEEYIITIGNHLATEEKFKSAKAAQ 40
Lambda K H a alpha
0.311 0.126 0.318 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4724881