bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
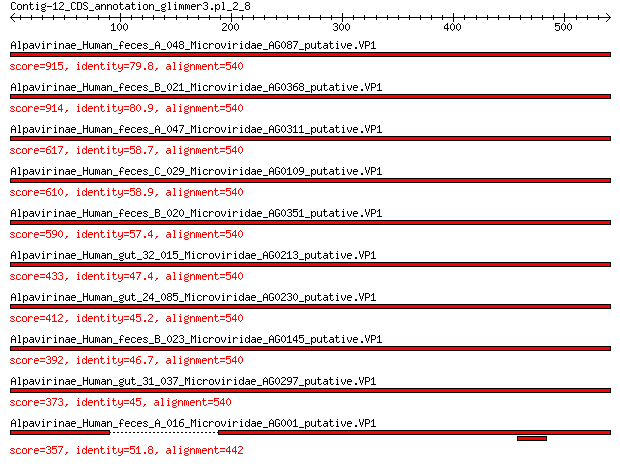
Query= Contig-12_CDS_annotation_glimmer3.pl_2_8
Length=540
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1 915 0.0
Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1 914 0.0
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 617 0.0
Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1 610 0.0
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 590 0.0
Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1 433 6e-146
Alpavirinae_Human_gut_24_085_Microviridae_AG0230_putative.VP1 412 9e-138
Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1 392 8e-130
Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1 373 2e-122
Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1 357 3e-116
> Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1
Length=650
Score = 915 bits (2365), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 431/542 (80%), Positives = 486/542 (90%), Gaps = 2/542 (0%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQE-VTVQKM 59
MSDIKLPQ+ ++INK DNPTP+NQW+Q+NPSCLLAYLGIRG+ N TT E VT+QKM
Sbjct 109 MSDIKLPQIRVNINKRNDNPTPENQWTQVNPSCLLAYLGIRGFGINALTTPNENVTIQKM 168
Query 60 AVPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASG 119
AVPL+GY+DIFKN+YANTQEENFYIIGAT I ++V Q G S TPDKIN GIA+G
Sbjct 169 AVPLIGYYDIFKNFYANTQEENFYIIGATTAIDKVEVAQKNGNQIISNTPDKINAGIANG 228
Query 120 DSVEITPKNTYEADELIITWFDMSTETTRTGKPTEFGTWSKTSGNWNVVMAPTQVGLLMS 179
D + I P NTYE+ EL +TWFD +++T+RTGKPTEFGTW+K +G W+V++ T VG LMS
Sbjct 229 DIITIYPINTYESSELAVTWFDAASQTSRTGKPTEFGTWNKATGKWSVIIPETTVGALMS 288
Query 180 ITPKNRVQLQDYPLEEIDEMRDKILSTKGNIT-SIPNNSNIALYDSFGKRLPNGKLNTTS 238
ITP NRVQL+ YPLE+ID +RD IL+TKGNI ++ N++I+L SF +RLPNGKLNT+S
Sbjct 289 ITPNNRVQLKTYPLEDIDTLRDNILTTKGNIVFNVTGNNSISLLKSFAERLPNGKLNTSS 348
Query 239 SQYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNMLNR 298
QYGLCLKTYNSDLLQNWINTEWIDGV GI+E++A+DVTDGKLTMDALNL QK+YNMLNR
Sbjct 349 VQYGLCLKTYNSDLLQNWINTEWIDGVAGINEISAVDVTDGKLTMDALNLSQKIYNMLNR 408
Query 299 IAVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTLAGR 358
IAVSGGTYRDWLETVFTGGNYMERCETP+FEGGMSTE+VFQEVISNSASGEQPLGTLAGR
Sbjct 409 IAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEVVFQEVISNSASGEQPLGTLAGR 468
Query 359 GYDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDGIGY 418
GYDTGKQKGGH+KIKVTEP FIMGIGSITPRIDYSQGNEF+NE T+DDIHKPALDGIGY
Sbjct 469 GYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFFNELQTVDDIHKPALDGIGY 528
Query 419 QDSLNWQRAWWDDNRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMVLNRN 478
QDSLNWQRAWWDD RM+ NGR+Q SAGKTVAW+NYMTNINRTFGNFAINDNEAFMVLNRN
Sbjct 529 QDSLNWQRAWWDDTRMQDNGRIQSSAGKTVAWINYMTNINRTFGNFAINDNEAFMVLNRN 588
Query 479 YEMSPSAGTNETKIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAKQIP 538
YE++P+AGTNETKIADLTTYIDPVK+NYIFA+TNLDAMNFWVQTKFDIKVRRLISAKQIP
Sbjct 589 YELNPNAGTNETKIADLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKVRRLISAKQIP 648
Query 539 NL 540
NL
Sbjct 649 NL 650
> Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1
Length=649
Score = 914 bits (2363), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 437/541 (81%), Positives = 483/541 (89%), Gaps = 1/541 (0%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
MSDIKLPQ+ +SINKT D PTP+NQWSQ+NPSCLLAYLGI+G+ N TT++ VTV+KMA
Sbjct 109 MSDIKLPQIKVSINKTIDLPTPENQWSQVNPSCLLAYLGIKGFGINATTSNAPVTVEKMA 168
Query 61 VPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASGD 120
VPL+GY+DIFKN+YANTQEENFYIIGAT I I V Q G STTPDKINIGIA+ D
Sbjct 169 VPLIGYYDIFKNFYANTQEENFYIIGATNAIYKITVGQTTGTKIESTTPDKINIGIANKD 228
Query 121 SVEITPKNTYEADELIITWFDMSTETTRTGKPTEFGTWSKTSGNWNVVMAPTQVGLLMSI 180
+V+I P NTYEADEL +TWFD +T+TT+TGKPT FGTW+K++G W V M T VG+LMSI
Sbjct 229 TVQIFPLNTYEADELTVTWFDAATQTTKTGKPTNFGTWNKSTGQWIVKMPETTVGVLMSI 288
Query 181 TPKNRVQLQDYPLEEIDEMRDKILSTKGNIT-SIPNNSNIALYDSFGKRLPNGKLNTTSS 239
TP +RV+L+ YPLE+ID +RD IL+TKGNIT ++ + +I L SF RLPN KLNT S
Sbjct 289 TPNSRVKLKTYPLEDIDTLRDNILTTKGNITFNVTGDESIPLLKSFAGRLPNSKLNTASV 348
Query 240 QYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNMLNRI 299
QYGLCLKTYNSDLLQNWINTEWIDGV GI+E++A+DVTDGKLTMDALNL QKVYNMLNRI
Sbjct 349 QYGLCLKTYNSDLLQNWINTEWIDGVTGINEISAVDVTDGKLTMDALNLAQKVYNMLNRI 408
Query 300 AVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTLAGRG 359
AVSGGTYRDWLETVFTGGNYMERCETP+FEGGMSTEIVFQEVISNSASGEQPLGTLAGRG
Sbjct 409 AVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEIVFQEVISNSASGEQPLGTLAGRG 468
Query 360 YDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDGIGYQ 419
YDTGKQKGGH+KIKVTEP FIMGIGSITPRIDYSQGNEFYNE T+DDIHKPALDGIGYQ
Sbjct 469 YDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFYNELKTVDDIHKPALDGIGYQ 528
Query 420 DSLNWQRAWWDDNRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMVLNRNY 479
DSLNWQRAWWDD RME NGR+Q SAGKTVAW+NYMTNINRTFGNFAINDNEAFMVLNRNY
Sbjct 529 DSLNWQRAWWDDTRMENNGRIQSSAGKTVAWINYMTNINRTFGNFAINDNEAFMVLNRNY 588
Query 480 EMSPSAGTNETKIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAKQIPN 539
E++P+AGTNETKIADLTTYIDPVK+NYIFA NLDAMNFWVQTKFDIKVRRLISAKQIPN
Sbjct 589 ELNPNAGTNETKIADLTTYIDPVKFNYIFAYKNLDAMNFWVQTKFDIKVRRLISAKQIPN 648
Query 540 L 540
L
Sbjct 649 L 649
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 617 bits (1592), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 317/548 (58%), Positives = 407/548 (74%), Gaps = 25/548 (5%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
MS +K+PQL++ + K DNPT DN+W+Q+NPSCLLAYLGIRGY T+++ + K A
Sbjct 107 MSQVKIPQLNVEL-KNTDNPTTDNEWTQVNPSCLLAYLGIRGYGSLIGTSAK--YIGKNA 163
Query 61 VPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASGD 120
VPL+GY+DIFKNYYANTQEE FY IG + + VT I S NI +++GD
Sbjct 164 VPLIGYYDIFKNYYANTQEEKFYTIGG---VLPLNVTYEGTEIPNSGN----NIALSNGD 216
Query 121 SVEITPKNTYEADELIITWFDMSTETTRTG--KPTEFGTWSKTSGNWNVVMAPTQVGLLM 178
++ + N +++++ + +E R KP++ GT + + N + ++A + +L
Sbjct 217 TMVFS--NVTNINDVVL----IGSENLRYREWKPSDLGTVTGLTLNIDKLLAGNEY-ILH 269
Query 179 SITPKNRVQLQDYPLEEIDEMRDKILSTKGNIT-SIPNNSNIALYDSFGKRL---PNGKL 234
+ K V L + LE +D +RD IL+TKGN+ ++ ++ + F +R G L
Sbjct 270 KMYKKGMVDLNQWELESLDTLRDNILTTKGNVAFNVTGKDSVPVLKQFNERFGEKQTGYL 329
Query 235 NTTSSQYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYN 294
TT+ Q+GL LKTYNSDL QNWINT+WI+GVNGI+E++++DV+DG L+MDALNL QKVYN
Sbjct 330 KTTAPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGSLSMDALNLAQKVYN 389
Query 295 MLNRIAVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGT 354
MLNRIAVSGGTYRDWLETVFTGG YMERCETPIFEGG S EI+FQEVISNSA+ ++PLGT
Sbjct 390 MLNRIAVSGGTYRDWLETVFTGGEYMERCETPIFEGGTSQEIIFQEVISNSATEQEPLGT 449
Query 355 LAGRGYDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALD 414
LAGRG T KQ+GGHVKIKVTEP +IM I SITPRIDYSQGN++ E T+DD+HKPALD
Sbjct 450 LAGRGITTNKQRGGHVKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPALD 509
Query 415 GIGYQDSLNWQRAWWDDNRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMV 474
GIGYQDS+N +RAWW + +++ +AGKTVAW+NYMTN+NRT+GNFAI DNEAFMV
Sbjct 510 GIGYQDSINSERAWWAGYYSQGPLKVETAAGKTVAWINYMTNVNRTYGNFAIKDNEAFMV 569
Query 475 LNRNYEMSPSAGTNET--KIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLI 532
+NRNYE+ + G T +I DL+TYIDPVK+NYIFA+T+L+AMNFWVQTKFDIK RRLI
Sbjct 570 MNRNYELQINGGPTPTSIRIGDLSTYIDPVKFNYIFADTSLEAMNFWVQTKFDIKARRLI 629
Query 533 SAKQIPNL 540
SAKQIPNL
Sbjct 630 SAKQIPNL 637
> Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1
Length=640
Score = 610 bits (1573), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 318/545 (58%), Positives = 394/545 (72%), Gaps = 16/545 (3%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
MS +K+PQL++ + T DNPT N+W+QINPSCLLAYLGIRGY T++ V+K A
Sbjct 107 MSQVKIPQLNVGLKNT-DNPTSKNEWTQINPSCLLAYLGIRGYGSLIGVTTK--YVEKNA 163
Query 61 VPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASGD 120
VPL+GY+DIFKNYYANTQE+ FY IG+ T I + I IN + GD
Sbjct 164 VPLIGYYDIFKNYYANTQEDKFYTIGSGRKFT-ITINNVSYPI------TDINKTLQVGD 216
Query 121 SVEITPKNTYEADELIITWFDMSTETTRTGKPTEFGTWSKTSGNWNVVMAPTQVGLLMSI 180
++ TP T E +I + R ++ GTW++ + + + L I
Sbjct 217 VIKFTPTPTKEEQGNLI-FIGSENSRYRRWSGSDIGTWTEQNDFKISGLKAGKRYELDRI 275
Query 181 TPKNRVQLQDYPLEEIDEMRDKILSTKGNIT-SIPNNSNIALYDSFGKRLP--NGKLNTT 237
+ V LQ + LE +D +RD IL+TKGNIT S+ ++ + F +RL + L T
Sbjct 276 FNELSVGLQQWELESLDTLRDNILTTKGNITFSVTGIDSVPVLQQFNERLGANSEHLKTI 335
Query 238 SSQYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNMLN 297
Q+GL LKTYNSDL QNWINT+WI+GVNGI+E++++DV+DG L+MDALNL QKVYNMLN
Sbjct 336 GPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGNLSMDALNLAQKVYNMLN 395
Query 298 RIAVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTLAG 357
RIAVSGGTYRDWLETVFTGG YMERCETP+FEGG S EIVFQEVISNSA+ ++PLGTLAG
Sbjct 396 RIAVSGGTYRDWLETVFTGGEYMERCETPVFEGGTSQEIVFQEVISNSATEQEPLGTLAG 455
Query 358 RGYDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDGIG 417
RG T KQ+GGH+KIKVTEP +IM I SITPRIDYSQGN++ E T+DD+HKPALDGIG
Sbjct 456 RGITTNKQRGGHIKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPALDGIG 515
Query 418 YQDSLNWQRAWWDDNRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMVLNR 477
YQDS+N +RAWW + + +AGKTVAW+NYMTN+NRT+GNFAI DNEAFMV+NR
Sbjct 516 YQDSINSERAWWAGYYNADPDKKETAAGKTVAWINYMTNVNRTYGNFAIKDNEAFMVMNR 575
Query 478 NYEMSPSAGTNETK--IADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAK 535
NYE+ + G+ TK I DL+TYIDP K+NYIFA+T+L+AMNFWVQTKFDIK RRLISAK
Sbjct 576 NYELEITGGSTPTKITIGDLSTYIDPAKFNYIFADTSLEAMNFWVQTKFDIKARRLISAK 635
Query 536 QIPNL 540
QIPNL
Sbjct 636 QIPNL 640
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 590 bits (1520), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 310/547 (57%), Positives = 383/547 (70%), Gaps = 11/547 (2%)
Query 1 MSDIKLPQLSISINKTKDNPTPD-NQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKM 59
MS IKLPQLS+++NK +D P D QWSQINPSCLL+YLGI+GY + QK
Sbjct 109 MSQIKLPQLSVTLNKLQDQPGEDEQQWSQINPSCLLSYLGIKGYG-GIHKAAPTTEAQKN 167
Query 60 AVPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASG 119
AV +LGY+DIFKNYYAN QEEN+Y+IG+ + ++ IK+ I + P I SG
Sbjct 168 AVKVLGYWDIFKNYYANKQEENYYMIGSNDPLS-IKINNVT-IIDPNNIPSNIGTIKKSG 225
Query 120 DSVEITPKNTYEADE--LIITWFDMSTETTRTGKPTEFGTWSKTSGNWNVVMAPTQVGLL 177
P Y L +T + T G W + P
Sbjct 226 TIAIADPLKIYNTSNVTLWVTQQIGHSPVKMTVNEVGQGVWEENEFVITANNVPENSSWW 285
Query 178 MSITPKNRVQ-LQDYPLEEIDEMRDKILSTKGNITSI-PNNSNIALYDSFGKRLPNGKLN 235
+ I + Q L +PL+E D +RD+IL+ KGN I NS+ + F +R N KL
Sbjct 286 IRIIQSTQQQALVSFPLDEFDTLRDEILAKKGNQAFILSKNSSTKIAKIFNERGSNNKLK 345
Query 236 TTSSQYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNM 295
TT QYGL LKTYNSDL QNWINT+WIDG NGI+E++++DV++GKLTMDALNL QKVYNM
Sbjct 346 TTRPQYGLLLKTYNSDLYQNWINTDWIDGANGINEISSVDVSEGKLTMDALNLAQKVYNM 405
Query 296 LNRIAVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTL 355
LNRIAVSGGTYRDWLETV+ G Y+ERCETP FEGG S EIVFQEV+SNSA+ ++PLGTL
Sbjct 406 LNRIAVSGGTYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATEDEPLGTL 465
Query 356 AGRGYDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDG 415
AGRG + GKQKGG +K++ TEP +IM I SITPRIDYSQGN+F ++ TLDD+HKPALDG
Sbjct 466 AGRGVNAGKQKGGKIKVRATEPGYIMCITSITPRIDYSQGNDFDTDWKTLDDMHKPALDG 525
Query 416 IGYQDSLNWQRAWWDD--NRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFM 473
IGYQDS+N RAWWDD ++ ++ +AGKTVAW++YMTN+N+T+GNFA +EAFM
Sbjct 526 IGYQDSVNTGRAWWDDVYTGAQETNLVKHTAGKTVAWIDYMTNVNKTYGNFAAGMSEAFM 585
Query 474 VLNRNYEMSPSAGTNETKIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLIS 533
VLNRNYE+ +GTN +IADLTTYIDP+KYNYIFA+T++DAMNFWVQ KFDI RRL+S
Sbjct 586 VLNRNYEIKYDSGTNP-RIADLTTYIDPIKYNYIFADTSIDAMNFWVQIKFDITARRLMS 644
Query 534 AKQIPNL 540
AKQIPNL
Sbjct 645 AKQIPNL 651
> Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1
Length=653
Score = 433 bits (1114), Expect = 6e-146, Method: Compositional matrix adjust.
Identities = 256/572 (45%), Positives = 342/572 (60%), Gaps = 57/572 (10%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
M D+KLP ++S D+ N + N S L YLGI+G + V K A
Sbjct 107 MKDVKLPTFTLSGTGGGDS-NRGNFEKKTNSSSLPFYLGIKG-------ARGKKVVDKAA 158
Query 61 ---VPLLGYFDIFKNYYANTQEENFYI-IGATETITNIKVTQAEGAIHTSTTPDKI---- 112
PLL Y+DIFKNY+ANTQEE+FY+ IG E +K T A S T DK+
Sbjct 159 FQGTPLLMYYDIFKNYFANTQEESFYVAIGVGEDPLILKATWA------SNTKDKVFYSE 212
Query 113 ------NIGIASGDSVEITPKNTYEADELIITWFDMSTETTRTGKP------TEFGTWSK 160
NI + + I K A + +I + T T TG + GTW+
Sbjct 213 QDYSAENITVEGEMVLSIEDKTLTSAQKCLIFQNMLLTYKTATGGSQTSRSMSSLGTWNN 272
Query 161 T----------SGNWNVVMAPTQVGLLMSITPKNRVQLQDYPLEEIDEMRDKILSTKGN- 209
+G W M +L K ++L+ Y LEE++ ++D+IL+TKG+
Sbjct 273 ADETFVINKVLTGKWYPCMFSPDKTML-----KRVIKLKKYKLEELETIKDRILATKGDT 327
Query 210 ITSIPNNSNIALYDSFGKRLPNGKLNTTSSQYGLCLKTYNSDLLQNWINTEWIDGVNGIS 269
+ I + S G L ++ GL +KTY+SD+ QNW+NTE IDG GI+
Sbjct 328 VLEITDESEET---GVGTSLFKDIATNPTTMAGLAVKTYDSDVFQNWVNTELIDGAKGIN 384
Query 270 EVTAIDVTDGKLTMDALNLQQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPIFE 329
E +++ + D KL+MDALNL QKVY+ LNRIAVSG TY+DWLET +T GNY+ER ETP+FE
Sbjct 385 EASSVAIVDNKLSMDALNLAQKVYDFLNRIAVSGNTYKDWLETAYTAGNYIERPETPLFE 444
Query 330 GGMSTEIVFQEVISNSASGEQPLGTLAGRGYDTGKQKGGHVKIKVTEPSFIMGIGSITPR 389
GGM+ I FQEVISN+ + ++PLGTLAGRG T ++ G + +K++EPS+I+GI +ITP
Sbjct 445 GGMTQLIEFQEVISNAGTEQEPLGTLAGRGVTTQQRGDGEIYMKISEPSYIIGIVAITPM 504
Query 390 IDYSQGNEF-YNEFLTLDDIHKPALDGIGYQDSLNWQRAWWDDNRMEKNGRLQPSAGKTV 448
IDYSQGN++ +DD HKPA DGIG++DS+N QRA+W + AGKTV
Sbjct 505 IDYSQGNDWDMTNIKNMDDFHKPAFDGIGFEDSMNEQRAYWTAEYSDGEKISDTKAGKTV 564
Query 449 AWLNYMTNINRTFGNFAINDNEAFMVLNRNYEMSPSAGTNETKIADLTTYIDPVKYNYIF 508
AW+NYMTN N+TFG FA ++E FMV+NRNYE +++ I+DL+TYIDP KYN IF
Sbjct 565 AWINYMTNFNKTFGEFAAGESEDFMVMNRNYERDED---DDSLISDLSTYIDPSKYNQIF 621
Query 509 AETNLDAMNFWVQTKFDIKVRRLISAKQIPNL 540
A+ +L A NFWVQT I+VRR ISAKQIPNL
Sbjct 622 ADVSLSAQNFWVQTAAQIEVRRNISAKQIPNL 653
> Alpavirinae_Human_gut_24_085_Microviridae_AG0230_putative.VP1
Length=653
Score = 412 bits (1060), Expect = 9e-138, Method: Compositional matrix adjust.
Identities = 244/564 (43%), Positives = 341/564 (60%), Gaps = 41/564 (7%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
M ++KLP ++S D+ + + + NPS L YLGI+G T E
Sbjct 107 MKEVKLPTFTLSGTGGGDSKRGNFEM-KTNPSSLPFYLGIKGARGKET----EDRAAFQG 161
Query 61 VPLLGYFDIFKNYYANTQEENFYII-GATETITNIKVTQAEG----AIHTSTTPDKINIG 115
PLL YFDIFKNY+ANTQEE+FY+ G E +K T + ++ + NI
Sbjct 162 TPLLMYFDIFKNYFANTQEESFYVANGVGEDPLILKATWSNNTENKVFYSEQDYNAENIL 221
Query 116 IASGDSVEITPKNTYEADE------LIITWFDMSTETTRTGKPTE-FGTWSKT------- 161
+ + I K A + +I+T + ++T +T + + GTW+
Sbjct 222 VTGEMVLSIEDKTLTSAQKSLIFQNMILT-YKITTGGIQTSRSMDTLGTWNNADETFVID 280
Query 162 ---SGNWNVVMAPTQVGLLMSITPKNRVQLQDYPLEEIDEMRDKILSTKGN-ITSIPNNS 217
+GNW M +L I + +L+D ++ ++D+IL+TKG+ + I + S
Sbjct 281 KVLTGNWYPCMFSPDKTMLKKIIKLKKYKLED-----LETLKDRILATKGDTVLEITDES 335
Query 218 NIALYDSFGKRLPNGKLNTTSSQYGLCLKTYNSDLLQNWINTEWIDGVNGISEVTAIDVT 277
+ G L ++ GL +KTY+SD+ QNW+NTE I+G GI+E +++ +
Sbjct 336 DTT---GVGMSLFKDIAVNPTTMAGLAVKTYDSDVFQNWVNTELIEGAKGINEASSVAII 392
Query 278 DGKLTMDALNLQQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPIFEGGMSTEIV 337
D KL+MDALNL QKVY+ LNRIAVSG TY+DWLET +T GNY+ER ETP+FEGGM+ I
Sbjct 393 DNKLSMDALNLAQKVYDFLNRIAVSGNTYKDWLETAYTAGNYIERPETPLFEGGMTQLIE 452
Query 338 FQEVISNSASGEQPLGTLAGRGYDTGKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNE 397
FQEV+SN+ + ++PLG+LAGRG T ++ G + +K++EPS+IMGI +ITP IDYSQGN+
Sbjct 453 FQEVVSNAGTEQEPLGSLAGRGVTTQQKGDGEIYMKISEPSYIMGIVAITPMIDYSQGND 512
Query 398 F-YNEFLTLDDIHKPALDGIGYQDSLNWQRAWWDDNRMEKNGRLQPSAGKTVAWLNYMTN 456
+ +DD HKPA DGIG++DS+N QRA+W AGKTVAW+NYMTN
Sbjct 513 WDMTNIKNMDDFHKPAFDGIGFEDSMNEQRAYWTAEYNNGEKISDTKAGKTVAWINYMTN 572
Query 457 INRTFGNFAINDNEAFMVLNRNYEMSPSAGTNETKIADLTTYIDPVKYNYIFAETNLDAM 516
N+TFG FA ++E FMV+NRNYE N++ I+DL+TYIDP KYN IFA+ +L A
Sbjct 573 FNKTFGEFAAGESEDFMVMNRNYERDEE---NDSLISDLSTYIDPSKYNQIFADESLSAQ 629
Query 517 NFWVQTKFDIKVRRLISAKQIPNL 540
NFWVQT ++VRR ISAKQIPNL
Sbjct 630 NFWVQTAVQMEVRRNISAKQIPNL 653
> Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1
Length=668
Score = 392 bits (1007), Expect = 8e-130, Method: Compositional matrix adjust.
Identities = 252/589 (43%), Positives = 332/589 (56%), Gaps = 78/589 (13%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
MS +K+P + K N + D +S SC+ YLG RG +TS T A
Sbjct 109 MSKVKIPIYRVG-QKQGTNLSKDLLYSS---SCIFTYLGNRGQETKKNSTSNVNTQDFNA 164
Query 61 VPLLGYFDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIHTSTTPDKINIGIASGD 120
+PLLGY DIFKNYYAN QEE F IGA + I T ++ +G
Sbjct 165 IPLLGYLDIFKNYYANKQEEKFKFIGADPYLIINTANLKNNPISTGVGSQRV-----TGG 219
Query 121 SVEITPKN------TYEADELIITWFD-MSTETT-----RTGKPTEFGTW---------- 158
+ IT +N D++ IT+ + TET RT TE G W
Sbjct 220 TFNITLQNGPSDWKEVNKDDIEITFTQKIGTETEYQTVERTYTLTELGKWFNSTPILTIL 279
Query 159 ------SKTSGNWNVVMAPTQVGLLMSITPKNRVQLQDYPLEEIDEMRDKILSTKGN--- 209
S T+G+ +++ G+ S KN + L ++D++R+ IL+ KGN
Sbjct 280 TLAYNNSGTTGSQDLLY-----GIPTSYKIKNITNILTENLSDLDQLREDILA-KGNERF 333
Query 210 ----ITSIPNNSNIALYDSFGKRLPNG---------KLNTTSSQYGLCLKTYNSDLLQNW 256
T + + N ++ + + G K N +S Q GL LKTY SD+ NW
Sbjct 334 EMNLTTKLGGSINF----TYIQHILGGNESSYEKATKYNRSSPQCGLLLKTYQSDIFTNW 389
Query 257 INTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNMLNRIAVSGGTYRDWLETVFTG 316
IN+EWIDG NGIS +TAI K T+D LNL +KVY+MLNRIA+SGGTY+DW+ETV+T
Sbjct 390 INSEWIDGENGISAITAISTEGNKFTIDQLNLSKKVYDMLNRIAISGGTYQDWVETVYT- 448
Query 317 GNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTLAGRGYDTGKQKGGHVKIKVTE 376
+ ETP++EGGMS EI FQEV+SNSA+ E+PLGTLAGRG+ + K KGG + IKVTE
Sbjct 449 SEWNMHTETPVYEGGMSAEIEFQEVVSNSATEEEPLGTLAGRGFASNK-KGGQLHIKVTE 507
Query 377 PSFIMGIGSITPRIDYSQGNEF-YNEFLTLDDIHKPALDGIGYQDSLNWQRAWWDDNRME 435
P +IMGI SITPR+DY QGN++ T+DDIHKP LD IGYQD + Q M
Sbjct 508 PCYIMGIASITPRVDYCQGNDWDITSLDTMDDIHKPQLDSIGYQDLMQEQ--------MN 559
Query 436 KNGRLQPSAGKTVAWLNYMTNINRTFGNFAIND--NEAFMVLNRNYEMSPSAGTNET--K 491
+ GK +W+NYMT+ N+T+G FA D EAFMVLNR +++ ET K
Sbjct 560 AQASRNLAVGKQPSWINYMTSFNKTYGTFANEDGEGEAFMVLNRYFDIKEIDAGTETGVK 619
Query 492 IADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAKQIPNL 540
+ + +TYIDP +YNYIFAET +MNFWVQ F I+ RR++SA QIPNL
Sbjct 620 VYNTSTYIDPSQYNYIFAETGTKSMNFWVQLGFGIEARRVMSASQIPNL 668
> Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1
Length=668
Score = 373 bits (958), Expect = 2e-122, Method: Compositional matrix adjust.
Identities = 243/591 (41%), Positives = 331/591 (56%), Gaps = 81/591 (14%)
Query 1 MSDIKLPQLSISINK------TKDNPTPDNQWSQINPSCLLAYLGIRGYAY-NTTTTSQE 53
MS IKLP+++++ K KDNP + PS LL YLGI N + E
Sbjct 108 MSQIKLPKVTLTAYKQYATDVKKDNP--------LQPSSLLTYLGISNVGMINGSDPFAE 159
Query 54 VTVQKMAVPLLGYFDIFKNYYANTQEENFYIIGATETITNIK-----VTQAEGAIHTSTT 108
AVP+L Y+DIFKNYYAN QE+ FY+ ++ + ++ + T
Sbjct 160 RNFN--AVPMLAYYDIFKNYYANKQEDKFYVRNFKNIVSAVSGWSKLENGSQNRLWNELT 217
Query 109 PDKIN--IGIASGDSVEITPKNTYEADEL--------------------------IITWF 140
D+ N ++ ++IT D L +IT+F
Sbjct 218 ADRFNSAYSMSLNADIKITTSEAISKDNLEAKCEIYYGYVNNTPAENLYTFKWIPLITYF 277
Query 141 DMSTETTRTGKPTEFGTWSKTSGNWNVVMAPTQVGLLMSITPKNRVQLQDYPLEEIDEMR 200
D + + T T +T N +V +GL + I +N Q+ L++ID +R
Sbjct 278 DYISYDSATQTYTY--RVKRTEYNRTIVFL---MGLRIQIG-RNSAQI----LDDIDTIR 327
Query 201 DKILSTKGNITSIPNNS----------NIALYDSFGKRLPNGKLNTTSSQYGLCLKTYNS 250
+ IL+T GN I N N + S + NG S ++GL KTY S
Sbjct 328 EDILATPGNRNWIFQNREGQENEIPFMNFLMNQSSNNK--NGGGFLFSPEWGLVTKTYQS 385
Query 251 DLLQNWINTEWIDGVNGISEVTAIDVTDGKLTMDALNLQQKVYNMLNRIAVSGGTYRDWL 310
DL QNWINTEW+DG +GI+++TA+ V DGK T+DALNL KVYN LNRIA+S G+YR WL
Sbjct 386 DLCQNWINTEWLDGDDGINQITAVAVEDGKFTIDALNLANKVYNYLNRIAISDGSYRSWL 445
Query 311 ETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGEQPLGTLAGRGYDTGKQKGGHV 370
ET +T G+Y ER ETPI+ GG S EI+FQEV+S SA+ ++PLG+LAGRG D+ KGG+V
Sbjct 446 ETTWT-GSYTERTETPIYCGGSSAEIIFQEVVSTSAATDEPLGSLAGRGTDS-NHKGGYV 503
Query 371 KIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDGIGYQD-SLNWQRAWW 429
++ TEP +++GI SITPR+DY+QGN++ ++DD+HKPALDGIG+QD S A
Sbjct 504 TVRATEPGYLIGITSITPRLDYTQGNQWDVNLDSIDDLHKPALDGIGFQDLSAELLHAGT 563
Query 430 DDNRMEKNGRLQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMVLNRNYEMSPSAGTNE 489
+ Q GK AW++YMTNINR +GNF N+N FM+L+R Y + T
Sbjct 564 TQINTINDTITQKFIGKQPAWIDYMTNINRAYGNFRTNEN--FMILSRQYSLDYKKNT-- 619
Query 490 TKIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAKQIPNL 540
I D+TTYIDP YN IFA+ + DA NFWVQ +I+ RRL+SAK IPNL
Sbjct 620 --IKDMTTYIDPDLYNGIFADQSFDAQNFWVQIGIEIEARRLMSAKIIPNL 668
> Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1
Length=657
Score = 357 bits (915), Expect = 3e-116, Method: Compositional matrix adjust.
Identities = 189/360 (53%), Positives = 247/360 (69%), Gaps = 21/360 (6%)
Query 188 LQDYPLEEIDEMRDKILSTKGN-ITSIPNNSNIALYDSFG--KRLPNGKLNTTSSQYGLC 244
L++ LE +D++RD+IL GN I + +N + + F ++ KL GL
Sbjct 312 LKEEKLEVLDKIRDEILKNPGNNILQLQTTTNTDVNNLFEDIRKASKNKLG------GLV 365
Query 245 LKTYNSDLLQNWINTEWIDGVNGISEVTAIDVT-DGKLTMDALNLQQKVYNMLNRIAVSG 303
+KTY+SD+ NW+ TEWIDG NGIS+VTA+ DG +TMDALNLQQKVYNMLNRIAVSG
Sbjct 366 IKTYDSDIFNNWVKTEWIDGENGISKVTALKPNEDGTITMDALNLQQKVYNMLNRIAVSG 425
Query 304 GTYRDWLETVFTGGNYMERCETPIFEGGMSTEIVFQEVISNSASGE-QPLGTLAGRGYDT 362
GTYRDWLETV+T G Y++R ETP+F+GGMS I F EV++ +++ E Q LG LAGRGY
Sbjct 426 GTYRDWLETVYTAGRYLDRPETPVFQGGMSQMIEFDEVVATTSNNEGQALGELAGRGYAR 485
Query 363 GKQKGGHVKIKVTEPSFIMGIGSITPRIDYSQGNEFYNEFLTLDDIHKPALDGIGYQDSL 422
G + +V EP ++MG+ +ITP +DYSQGN+F T+DD+HKPALDGIGYQD +
Sbjct 486 QPNSSGRLHFQVEEPGYVMGLVAITPMVDYSQGNDFDLNLFTMDDLHKPALDGIGYQDLM 545
Query 423 NWQRAWWDDNRMEKNGR--LQPSAGKTVAWLNYMTNINRTFGNFAINDNEAFMVLNRNYE 480
N QRAWW +NG + GK+VAW++YMTN NRTFGNFA ++E FMVLNRNYE
Sbjct 546 NEQRAWW---TATQNGTKITDTTPGKSVAWIDYMTNFNRTFGNFATGESEDFMVLNRNYE 602
Query 481 MSPSAGTNETKIADLTTYIDPVKYNYIFAETNLDAMNFWVQTKFDIKVRRLISAKQIPNL 540
+ G I++ +TYI+P ++ IFA+T LD+ NFW+QT +I R SAKQIPN
Sbjct 603 DDEADG-----ISNGSTYINPQEHIDIFADTALDSQNFWIQTACEITRRGNYSAKQIPNF 657
Score = 66.2 bits (160), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 40/90 (44%), Positives = 52/90 (58%), Gaps = 11/90 (12%)
Query 1 MSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQKMA 60
MSDIK P + + N + NPS L +YLG +G T ++ + K A
Sbjct 109 MSDIKFPTIY------RGNELDSKNKIKYNPSSLYSYLGWKG----TKQSATGIGAMKQA 158
Query 61 VPLLGYFDIFKNYYANTQEENFYII-GATE 89
VP L Y DIFKNY+ANTQE+NFYII G+T+
Sbjct 159 VPWLIYLDIFKNYFANTQEDNFYIITGSTK 188
Score = 22.7 bits (47), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 13/26 (50%), Gaps = 0/26 (0%)
Query 457 INRTFGNFAINDNEAFMVLNRNYEMS 482
I RT G + DN V R Y+MS
Sbjct 3 ITRTIGKNTLGDNNKMKVRLREYDMS 28
Lambda K H a alpha
0.315 0.132 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 49252622