bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_8
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
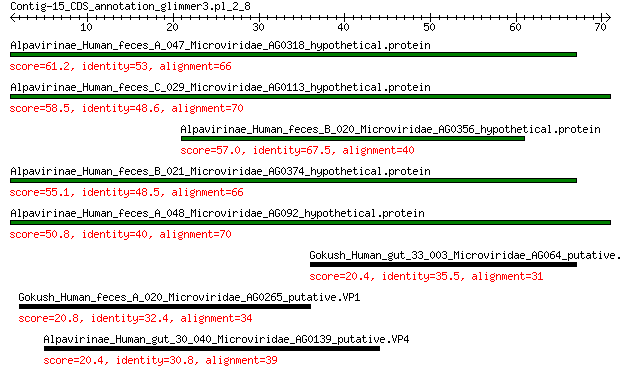
Alpavirinae_Human_feces_A_047_Microviridae_AG0318_hypothetical.... 61.2 1e-15
Alpavirinae_Human_feces_C_029_Microviridae_AG0113_hypothetical.... 58.5 1e-14
Alpavirinae_Human_feces_B_020_Microviridae_AG0356_hypothetical.... 57.0 4e-14
Alpavirinae_Human_feces_B_021_Microviridae_AG0374_hypothetical.... 55.1 2e-13
Alpavirinae_Human_feces_A_048_Microviridae_AG092_hypothetical.p... 50.8 1e-11
Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M... 20.4 1.9
Gokush_Human_feces_A_020_Microviridae_AG0265_putative.VP1 20.8 2.1
Alpavirinae_Human_gut_30_040_Microviridae_AG0139_putative.VP4 20.4 2.7
> Alpavirinae_Human_feces_A_047_Microviridae_AG0318_hypothetical.protein
Length=66
Score = 61.2 bits (147), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 45/67 (67%), Gaps = 3/67 (4%)
Query 1 MAATKFSVITTGNNPSFHMSIKELPAECTYKIAVTIAKAI-VPKDERIIAVVESWKLYPN 59
MAATKF+ I NN + +E+P TYKIA AK + P++ +++ VVESWKLYP
Sbjct 1 MAATKFTAIYADNNGK--IIEREIPGMNTYKIAEKFAKMLNDPEETKLVCVVESWKLYPK 58
Query 60 ENEKTEK 66
ENEKTEK
Sbjct 59 ENEKTEK 65
> Alpavirinae_Human_feces_C_029_Microviridae_AG0113_hypothetical.protein
Length=73
Score = 58.5 bits (140), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/71 (48%), Positives = 45/71 (63%), Gaps = 3/71 (4%)
Query 1 MAATKFSVITTGNNPSFHMSIKELPAECTYKIAVTIAKAI-VPKDERIIAVVESWKLYPN 59
MAATKF+ I N + +E+P TYKIA A + PK+ +++ VVESWKLYP
Sbjct 1 MAATKFTAIYVNNEGK--IIEREIPGMNTYKIAEKFAMMLNDPKETKLVCVVESWKLYPK 58
Query 60 ENEKTEKKQKI 70
ENEKT+KK +
Sbjct 59 ENEKTKKKDNV 69
> Alpavirinae_Human_feces_B_020_Microviridae_AG0356_hypothetical.protein
Length=55
Score = 57.0 bits (136), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 27/42 (64%), Positives = 35/42 (83%), Gaps = 2/42 (5%)
Query 21 IKELPAECTYKIAVTIAKAIVPK--DERIIAVVESWKLYPNE 60
+KELP + TYK+AVT+AKA++ + E I+AVVESWKLYPNE
Sbjct 1 MKELPGKHTYKVAVTVAKALMQEVEGEEIVAVVESWKLYPNE 42
> Alpavirinae_Human_feces_B_021_Microviridae_AG0374_hypothetical.protein
Length=66
Score = 55.1 bits (131), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 32/67 (48%), Positives = 43/67 (64%), Gaps = 3/67 (4%)
Query 1 MAATKFSVITTGNNPSFHMSIKELPAECTYKIAVTIAKAI-VPKDERIIAVVESWKLYPN 59
MAATKF+ I N + +E+P TYKIA A + P++ +++ V+ESWKLYP
Sbjct 1 MAATKFTAIYVNNEGK--IIEREIPGMNTYKIAEKFAIMLNDPEETKLVCVIESWKLYPK 58
Query 60 ENEKTEK 66
ENEKTEK
Sbjct 59 ENEKTEK 65
> Alpavirinae_Human_feces_A_048_Microviridae_AG092_hypothetical.protein
Length=73
Score = 50.8 bits (120), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 44/71 (62%), Gaps = 3/71 (4%)
Query 1 MAATKFSVITTGNNPSFHMSIKELPAECTYKIAVTIAKAI-VPKDERIIAVVESWKLYPN 59
MAATKF+ I N + +E+P +YKIA A + P++ +++ V+E+WK+YPN
Sbjct 1 MAATKFTAIYINNEGK--IIKREIPGMNSYKIAEKFAMMLNDPEETKLVCVIETWKMYPN 58
Query 60 ENEKTEKKQKI 70
EKTEK+ +
Sbjct 59 NYEKTEKEDNL 69
> Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M15_3
Length=130
Score = 20.4 bits (41), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 16/31 (52%), Gaps = 1/31 (3%)
Query 36 IAKAIVPKDERIIAVVESWKLYPNENEKTEK 66
+ +IVP II V ++W + NEK K
Sbjct 99 VLSSIVPNSGGII-VYDNWVHFDTRNEKYRK 128
> Gokush_Human_feces_A_020_Microviridae_AG0265_putative.VP1
Length=558
Score = 20.8 bits (42), Expect = 2.1, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 16/34 (47%), Gaps = 0/34 (0%)
Query 2 AATKFSVITTGNNPSFHMSIKELPAECTYKIAVT 35
+A ++ GN P + KEL TYK +T
Sbjct 213 SAESVTIPMLGNAPIYGYKEKELKTPWTYKNPMT 246
> Alpavirinae_Human_gut_30_040_Microviridae_AG0139_putative.VP4
Length=376
Score = 20.4 bits (41), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 12/39 (31%), Positives = 19/39 (49%), Gaps = 0/39 (0%)
Query 5 KFSVITTGNNPSFHMSIKELPAECTYKIAVTIAKAIVPK 43
K+ T G+NP ++KE E + A+ AK + K
Sbjct 278 KYGRETMGDNPIAWNAMKERRKEEKQRRAIAEAKKLAEK 316
Lambda K H a alpha
0.313 0.126 0.355 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3733230