bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_7
Length=639
Score E
Sequences producing significant alignments: (Bits) Value
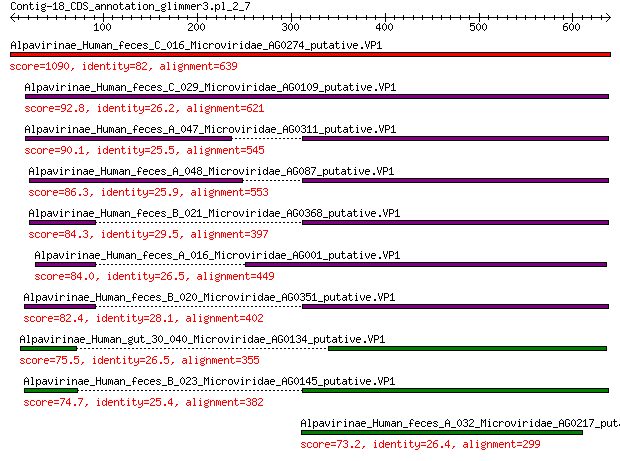
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 1090 0.0
Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1 92.8 2e-21
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 90.1 1e-20
Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1 86.3 2e-19
Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1 84.3 9e-19
Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1 84.0 9e-19
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 82.4 3e-18
Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1 75.5 5e-16
Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1 74.7 8e-16
Alpavirinae_Human_feces_A_032_Microviridae_AG0217_putative.VP1 73.2 3e-15
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 1090 bits (2818), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 524/641 (82%), Positives = 559/641 (87%), Gaps = 2/641 (0%)
Query 1 MPKYRIPSDYENQTPRAIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQP 60
M KYRIPSDY+NQTPRA+H R + +YGTIHPGLA+PVHHRHLNVGERIRGRID LLQSQP
Sbjct 1 MSKYRIPSDYQNQTPRAMHRRASSAYGTIHPGLAIPVHHRHLNVGERIRGRIDELLQSQP 60
Query 61 MLGPLMNGFKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPA 120
MLGPLMNGFKLITIATFTPDSAIYGWM NGRR+ PDEY KFGKVYF LAGPN+ NY DPA
Sbjct 61 MLGPLMNGFKLITIATFTPDSAIYGWMSNGRRFTPDEYMKFGKVYFSLAGPNLENYKDPA 120
Query 121 FQIARRVRRLTFGLELNADRAKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAV 180
F+++R VRRLTFGL+LN+D+ KIYESWVSDELN+TG GQPTHIGRGGLWDWLGIA GAV
Sbjct 121 FKVSRPVRRLTFGLDLNSDQKKIYESWVSDELNATGSSGQPTHIGRGGLWDWLGIAAGAV 180
Query 181 CPNLGIKLPSVA-GVRGQFYPPSFKFNGAPFFAYFLSHYYYIANMQEKYMYFTRGVGEMQ 239
CPNLG K S + GVRG+ YP SF+FN APF +YFLSHYYYIANMQE YMYFTRGVGEMQ
Sbjct 181 CPNLGKKTTSSSPGVRGEIYPSSFQFNAAPFISYFLSHYYYIANMQEDYMYFTRGVGEMQ 240
Query 240 RVRPDGQQESLYRPFFSDVFTSFKPDDFLLYLDTLSYRTRVGAEIDALTLQLEGIPI-NC 298
RVRPDGQQESLYRPFFSDVF S P+ FL LD + TR G + + +P N
Sbjct 241 RVRPDGQQESLYRPFFSDVFNSLDPNAFLYALDDVRNITRTGTGCELFKTFSDALPTSNS 300
Query 299 VQAMACAGIQGYGGLLSVPYSPDLFSNIIKQGSSPAVEIEVMNALDASTDTGFSVAVPEL 358
AMACAGIQGYGGLLSVPYSPDLF NIIKQGSSP EIEV+ ALD++TDTGFS+AVPEL
Sbjct 301 FVAMACAGIQGYGGLLSVPYSPDLFGNIIKQGSSPTAEIEVIKALDSNTDTGFSIAVPEL 360
Query 359 RLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMAN 418
RLKTK+QNWMDRLF+SGGRVGDVFRTLWG KSSAPYVNKPDFLGVWQASINPSNVRAMAN
Sbjct 361 RLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASINPSNVRAMAN 420
Query 419 GSASGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLA 478
GSASGEDANLGQLAACVDRYCDFS HSGIDYYAKEPGTFMLI MLVPEPAY QGLHPDLA
Sbjct 421 GSASGEDANLGQLAACVDRYCDFSDHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLA 480
Query 479 SISFGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMAS 538
SISFGDDFNPELNGIGFQ VPRHRF+MMPRG + Q+ +PW G AGTGV IDPN S
Sbjct 481 SISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNLTGLDQQTSPWFGNAGTGVVIDPNTVS 540
Query 539 VGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINP 598
VGEEVAWSWLRTDY RLHGDFAQNGNYQYWVL R FT+YFPDAGTGFY D E TYINP
Sbjct 541 VGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDAGTGFYLDGEYTGTYINP 600
Query 599 LDWQYVFVDQTLMAGNFAYYGYFDLKVTSSLSANYMPYLGR 639
LDWQYVFVDQTLMAGNF+YYG FDLKVTSSLSANYMPYLGR
Sbjct 601 LDWQYVFVDQTLMAGNFSYYGTFDLKVTSSLSANYMPYLGR 641
> Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1
Length=640
Score = 92.8 bits (229), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 163/696 (23%), Positives = 248/696 (36%), Gaps = 156/696 (22%)
Query 17 AIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIAT 76
+ H T PG VP + G+ I I++ + + P +GPL FKL
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHVY 85
Query 77 FTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIARRVRRLTFGLEL 136
P W+ N R TK G +D + Q+ ++ +L GL+
Sbjct 86 TVPFRLYNSWLHNNR-------TKIG--------------LDMS-QV--KIPQLNVGLK- 120
Query 137 NADRAKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAVCPNLGIK-LPSVAGVR 195
N D PT +W I P + LGI+ S+ GV
Sbjct 121 NTD--------------------NPTSKN-----EWTQINPSCLLAYLGIRGYGSLIGVT 155
Query 196 GQFYPPSFKFNGAPFFAYFLSHYYYIANMQEKYMYF-----------------------T 232
++ N P Y+ Y AN QE Y T
Sbjct 156 TKYVEK----NAVPLIGYYDIFKNYYANTQEDKFYTIGSGRKFTITINNVSYPITDINKT 211
Query 233 RGVGEMQRVRPD------------GQQESLYRPFF-SDVFTSFKPDDFLL---------Y 270
VG++ + P G + S YR + SD+ T + +DF +
Sbjct 212 LQVGDVIKFTPTPTKEEQGNLIFIGSENSRYRRWSGSDIGTWTEQNDFKISGLKAGKRYE 271
Query 271 LDTLSYRTRVGA---EIDAL-TLQ----------------LEGIPI----NCVQAMACAG 306
LD + VG E+++L TL+ ++ +P+ N
Sbjct 272 LDRIFNELSVGLQQWELESLDTLRDNILTTKGNITFSVTGIDSVPVLQQFNERLGANSEH 331
Query 307 IQGYG---GLLSVPYSPDLFSNIIKQGSSPAVE-IEVMNALDASTDTGFSVAVPELRLKT 362
++ G GL Y+ DL+ N I V I ++++D S ++++ L L
Sbjct 332 LKTIGPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDG---NLSMDALNLAQ 388
Query 363 KLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSAS 422
K+ N ++R+ VSGG D T++ P F G I V +++
Sbjct 389 KVYNMLNRIAVSGGTYRDWLETVFTGGEYMERCETPVFEGGTSQEI----VFQEVISNSA 444
Query 423 GEDANLGQLAA-CVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASIS 481
E LG LA + GH I EPG M I + P Y QG D +
Sbjct 445 TEQEPLGTLAGRGITTNKQRGGH--IKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKT 502
Query 482 FGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGE 541
D P L+GIG+Q + + E+ W GY D + G+
Sbjct 503 MDDLHKPALDGIGYQD----------------SINSERAWWAGYYNADP--DKKETAAGK 544
Query 542 EVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDW 601
VAW T+ R +G+FA N + V+ R + T STYI+P +
Sbjct 545 TVAWINYMTNVNRTYGNFAIKDNEAFMVMNRNYELEITGGSTPTKITIGDLSTYIDPAKF 604
Query 602 QYVFVDQTLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
Y+F D +L A NF FD+K +SA +P L
Sbjct 605 NYIFADTSLEAMNFWVQTKFDIKARRLISAKQIPNL 640
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 90.1 bits (222), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 90/329 (27%), Positives = 136/329 (41%), Gaps = 31/329 (9%)
Query 312 GLLSVPYSPDLFSNIIKQGSSPAVE-IEVMNALDASTDTGFSVAVPELRLKTKLQNWMDR 370
GL Y+ DL+ N I V I ++++D S S+++ L L K+ N ++R
Sbjct 337 GLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDG---SLSMDALNLAQKVYNMLNR 393
Query 371 LFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQ 430
+ VSGG D T++ P F G I V + +++ E LG
Sbjct 394 IAVSGGTYRDWLETVFTGGEYMERCETPIFEGGTSQEIIFQEVIS----NSATEQEPLGT 449
Query 431 LAA-CVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNPE 489
LA + GH I EPG M I + P Y QG D + D P
Sbjct 450 LAGRGITTNKQRGGHVKIK--VTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPA 507
Query 490 LNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTG-VTIDPNMASVGEEVAWSWL 548
L+GIG+Q + + E+ W GY G + ++ + G+ VAW
Sbjct 508 LDGIGYQD----------------SINSERAWWAGYYSQGPLKVE---TAAGKTVAWINY 548
Query 549 RTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQYVFVDQ 608
T+ R +G+FA N + V+ R + T STYI+P+ + Y+F D
Sbjct 549 MTNVNRTYGNFAIKDNEAFMVMNRNYELQINGGPTPTSIRIGDLSTYIDPVKFNYIFADT 608
Query 609 TLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
+L A NF FD+K +SA +P L
Sbjct 609 SLEAMNFWVQTKFDIKARRLISAKQIPNL 637
Score = 30.0 bits (66), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 49/220 (22%), Positives = 74/220 (34%), Gaps = 55/220 (25%)
Query 17 AIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIAT 76
+ H T PG VP + G+ I I++ + + P +GPL FKL
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHIY 85
Query 77 FTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIARRVRRLTFGLEL 136
P W+ N R TK G L
Sbjct 86 TIPFRLYNSWLHNNR-------TKIG---------------------------------L 105
Query 137 NADRAKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAVCPNLGIK-LPSVAGVR 195
+ + KI + ++ EL +T + PT +W + P + LGI+ S+ G
Sbjct 106 DMSQVKIPQ--LNVELKNTDN---PTTDN-----EWTQVNPSCLLAYLGIRGYGSLIGTS 155
Query 196 GQFYPPSFKFNGAPFFAYFLSHYYYIANMQEKYMYFTRGV 235
++ N P Y+ Y AN QE+ Y GV
Sbjct 156 AKYIGK----NAVPLIGYYDIFKNYYANTQEEKFYTIGGV 191
> Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1
Length=650
Score = 86.3 bits (212), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 98/331 (30%), Positives = 139/331 (42%), Gaps = 37/331 (11%)
Query 312 GLLSVPYSPDLFSNIIKQGSSPAVE-IEVMNALDASTDTGFSVAVPELRLKTKLQNWMDR 370
GL Y+ DL N I V I ++A+D T + + L L K+ N ++R
Sbjct 352 GLCLKTYNSDLLQNWINTEWIDGVAGINEISAVDV---TDGKLTMDALNLSQKIYNMLNR 408
Query 371 LFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQ 430
+ VSGG D T++ + P F G + + + + SASGE LG
Sbjct 409 IAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEVV---FQEVISNSASGEQP-LGT 464
Query 431 LAACVDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYFQG--LHPDLASISFGDDF 486
LA R D G I EP M I + P Y QG +L ++ D
Sbjct 465 LAG---RGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFFNELQTVD--DIH 519
Query 487 NPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEEVAWS 546
P L+GIG+Q + R D T Q+ +S G+ VAW
Sbjct 520 KPALDGIGYQDSLNWQ-----RAWWDDTRMQDNGRI-------------QSSAGKTVAWI 561
Query 547 WLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQYVFV 606
T+ R G+FA N N + VL R + P+AGT A+ +TYI+P+ + Y+F
Sbjct 562 NYMTNINRTFGNFAINDNEAFMVLNRNY-ELNPNAGTNETKIAD-LTTYIDPVKFNYIFA 619
Query 607 DQTLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
D L A NF FD+KV +SA +P L
Sbjct 620 DTNLDAMNFWVQTKFDIKVRRLISAKQIPNL 650
Score = 29.6 bits (65), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 45/230 (20%), Positives = 75/230 (33%), Gaps = 57/230 (25%)
Query 21 RVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIATFTPD 80
R S GT+ P L + G+ I I+S + + P +GPL FK P
Sbjct 37 RNTQSVGTLVPNLVLLAQK-----GDTIDIDIESHVLTHPTVGPLFGSFKHENHIFSVPL 91
Query 81 SAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIARRVRRLTFGLELNADR 140
W+ N R TK G LN
Sbjct 92 RLYNSWLHNNR-------TKIG---------------------------------LNMSD 111
Query 141 AKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAVCPNLGIKLPSVAGVRGQFYP 200
K+ + V+ +N D+ P + W + P + LGI+ + +
Sbjct 112 IKLPQIRVN--INKRNDNPTPEN-------QWTQVNPSCLLAYLGIRGFGINALTTPNEN 162
Query 201 PSFKFNGAPFFAYFLSHYYYIANMQEKYMYF---TRGVGEMQRVRPDGQQ 247
+ + P Y+ + AN QE+ Y T + +++ + +G Q
Sbjct 163 VTIQKMAVPLIGYYDIFKNFYANTQEENFYIIGATTAIDKVEVAQKNGNQ 212
> Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1
Length=649
Score = 84.3 bits (207), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 97/334 (29%), Positives = 138/334 (41%), Gaps = 43/334 (13%)
Query 312 GLLSVPYSPDLFSNIIK----QGSSPAVEIEVMNALDASTDTGFSVAVPELRLKTKLQNW 367
GL Y+ DL N I G + EI ++ D + + L L K+ N
Sbjct 351 GLCLKTYNSDLLQNWINTEWIDGVTGINEISAVDVTDGK------LTMDALNLAQKVYNM 404
Query 368 MDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDAN 427
++R+ VSGG D T++ + P F G I + + + SASGE
Sbjct 405 LNRIAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEIV---FQEVISNSASGEQP- 460
Query 428 LGQLAACVDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYFQG--LHPDLASISFG 483
LG LA R D G I EP M I + P Y QG + +L ++
Sbjct 461 LGTLAG---RGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFYNELKTVD-- 515
Query 484 DDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEEV 543
D P L+GIG+Q DD E N + +S G+ V
Sbjct 516 DIHKPALDGIGYQD----SLNWQRAWWDD--TRMENNGRI------------QSSAGKTV 557
Query 544 AWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQY 603
AW T+ R G+FA N N + VL R + P+AGT A+ +TYI+P+ + Y
Sbjct 558 AWINYMTNINRTFGNFAINDNEAFMVLNRNY-ELNPNAGTNETKIAD-LTTYIDPVKFNY 615
Query 604 VFVDQTLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
+F + L A NF FD+KV +SA +P L
Sbjct 616 IFAYKNLDAMNFWVQTKFDIKVRRLISAKQIPNL 649
Score = 28.9 bits (63), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 28/71 (39%), Gaps = 5/71 (7%)
Query 21 RVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIATFTPD 80
R S GT+ P L + G+ I I+S + + P +GPL FK P
Sbjct 37 RNTQSVGTLVPNLVLLAQK-----GDTIDIDIESHVLTHPTVGPLFGSFKHENHIFSVPI 91
Query 81 SAIYGWMRNGR 91
W+ N R
Sbjct 92 RLYNSWLHNNR 102
> Alpavirinae_Human_feces_A_016_Microviridae_AG001_putative.VP1
Length=657
Score = 84.0 bits (206), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 100/394 (25%), Positives = 162/394 (41%), Gaps = 47/394 (12%)
Query 251 YRPFFSDVFTSFKPDDFLLYLDTLSYRTRV----GAEIDALTLQLEGIPINCVQAMACAG 306
Y+ F + S ++ L LD + R + G I L N + + A
Sbjct 300 YKKMFQESLGSILKEEKLEVLDKI--RDEILKNPGNNILQLQTTTNTDVNNLFEDIRKAS 357
Query 307 IQGYGGLLSVPYSPDLFSNIIK-QGSSPAVEIEVMNALDASTDTGFSVAVPELRLKTKLQ 365
GGL+ Y D+F+N +K + I + AL + D ++ + L L+ K+
Sbjct 358 KNKLGGLVIKTYDSDIFNNWVKTEWIDGENGISKVTALKPNEDG--TITMDALNLQQKVY 415
Query 366 NWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPD---FLGVWQASINPSNVRAMANGSAS 422
N ++R+ VSGG D T++ ++ Y+++P+ F G I V A +++
Sbjct 416 NMLNRIAVSGGTYRDWLETVY---TAGRYLDRPETPVFQGGMSQMIEFDEVVAT---TSN 469
Query 423 GEDANLGQLAA-CVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASIS 481
E LG+LA R + SG + + +EPG M + + P Y QG DL +
Sbjct 470 NEGQALGELAGRGYARQPNSSGR--LHFQVEEPGYVMGLVAITPMVDYSQGNDFDLNLFT 527
Query 482 FGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGE 541
D P L+GIG+Q + E+ W G I + G+
Sbjct 528 MDDLHKPALDGIGYQDL-----------------MNEQRAWWTATQNGTKITD--TTPGK 568
Query 542 EVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDW 601
VAW T++ R G+FA + + VL R +Y D G + STYINP +
Sbjct 569 SVAWIDYMTNFNRTFGNFATGESEDFMVLNR---NYEDDEADGI----SNGSTYINPQEH 621
Query 602 QYVFVDQTLMAGNFAYYGYFDLKVTSSLSANYMP 635
+F D L + NF ++ + SA +P
Sbjct 622 IDIFADTALDSQNFWIQTACEITRRGNYSAKQIP 655
Score = 27.7 bits (60), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 28/65 (43%), Gaps = 2/65 (3%)
Query 28 TIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIATFTPDSAIYG-W 86
T+ G+ VP G+ ++ + + P LGPL FKL FT +Y W
Sbjct 39 TMGVGMLVPFMKILCQKGDIFDIKLTNKTMTHPTLGPLFGSFKLQHFI-FTAGFRLYNSW 97
Query 87 MRNGR 91
+ N R
Sbjct 98 LHNNR 102
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 82.4 bits (202), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 92/330 (28%), Positives = 135/330 (41%), Gaps = 34/330 (10%)
Query 312 GLLSVPYSPDLFSNIIKQG-SSPAVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDR 370
GLL Y+ DL+ N I A I ++++D S + + L L K+ N ++R
Sbjct 352 GLLLKTYNSDLYQNWINTDWIDGANGINEISSVDVSEG---KLTMDALNLAQKVYNMLNR 408
Query 371 LFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQ 430
+ VSGG D T++ P F G I V +++ ED LG
Sbjct 409 IAVSGGTYRDWLETVYASGQYIERCETPTFEGGTSQEI----VFQEVVSNSATEDEPLGT 464
Query 431 LAACVDRYCDFSGHSG--IDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNP 488
LA R + G I A EPG M I + P Y QG D + D P
Sbjct 465 LAG---RGVNAGKQKGGKIKVRATEPGYIMCITSITPRIDYSQGNDFDTDWKTLDDMHKP 521
Query 489 ELNGIGFQQVPRHRFTMMPRGMDDI-TAHQEKNPWLGYAGTGVTIDPNMASVGEEVAWSW 547
L+GIG+Q DD+ T QE N AG + VAW
Sbjct 522 ALDGIGYQD----SVNTGRAWWDDVYTGAQETNLVKHTAG-------------KTVAWID 564
Query 548 LRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQYVFVD 607
T+ + +G+FA + + VL R + + D+GT +TYI+P+ + Y+F D
Sbjct 565 YMTNVNKTYGNFAAGMSEAFMVLNRNYEIKY-DSGTN--PRIADLTTYIDPIKYNYIFAD 621
Query 608 QTLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
++ A NF FD+ +SA +P L
Sbjct 622 TSIDAMNFWVQIKFDITARRLMSAKQIPNL 651
Score = 33.9 bits (76), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/76 (28%), Positives = 29/76 (38%), Gaps = 0/76 (0%)
Query 16 RAIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIA 75
R+ H+ T PG VP + + I+S + + P GPL FKL
Sbjct 27 RSTHNLSTVIRNTQSPGTLVPTMTLVMQKDDTFDIEIESSVLTHPTTGPLYGSFKLENHL 86
Query 76 TFTPDSAIYGWMRNGR 91
F P W+ N R
Sbjct 87 FFIPFRLYNSWLHNNR 102
> Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1
Length=679
Score = 75.5 bits (184), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 77/298 (26%), Positives = 122/298 (41%), Gaps = 46/298 (15%)
Query 340 MNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPD 399
+N++ A + S + L L K+ ++R+ VS G +T++ ++ P
Sbjct 424 INSITAIDTSSGSFTLDTLNLAKKVYTMLNRIAVSDGSYNAWIQTVY-TSGGLNHIETPL 482
Query 400 FLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYCDFSGHSG--IDYYAKEPGTF 457
+LG I + + N S + ED LG LA + H G I + A EPG
Sbjct 483 YLGGSSLEIE---FQEVINNSGT-EDQPLGALAG----RGVATNHKGGNIVFKADEPGYI 534
Query 458 MLIAMLVPEPAYFQGLHPDLASISFGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQ 517
I + P YFQG DL S D P+L+GIGFQ + R ++ +
Sbjct 535 FCITSITPRVDYFQGNEWDLYLESLDDLHKPQLDGIGFQD-------RLYRHLNSSCVRE 587
Query 518 EKNPWLGYAGTGVTIDPNMASVGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSY 577
+ N S+G++ AW T+ + +G+FA N + L R F
Sbjct 588 DLN----------------ISIGKQPAWVQYMTNVNKTYGNFALVENEGWMCLNRIFE-- 629
Query 578 FPDAGTGFYGDAESYSTYINPLDWQYVFVDQTLMAGNFAYYGYFDLKVTSSLSANYMP 635
+ ++Y+TYI P + +F D L A NF F+ K +SA +P
Sbjct 630 ----------NTDTYTTYIQPHLYNNIFADTDLTAQNFWVQIAFNYKPRRVMSAKIIP 677
Score = 27.3 bits (59), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 17/59 (29%), Positives = 24/59 (41%), Gaps = 0/59 (0%)
Query 12 NQTPRAIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFK 70
N R+ H T+ PG+ VP L G+ +I + P +GPL FK
Sbjct 21 NNYNRSTHDNSFIMRTTMAPGVLVPTMTELLLPGDTYPIQIRCHTLTHPTIGPLFGSFK 79
> Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1
Length=668
Score = 74.7 bits (182), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 83/334 (25%), Positives = 146/334 (44%), Gaps = 48/334 (14%)
Query 312 GLLSVPYSPDLFSNIIKQGSSPAVEIEV-MNALDASTDTGFSVAVPELRLKTKLQNWMDR 370
GLL Y D+F+N I +S ++ E ++A+ A + G + +L L K+ + ++R
Sbjct 375 GLLLKTYQSDIFTNWI---NSEWIDGENGISAITAISTEGNKFTIDQLNLSKKVYDMLNR 431
Query 371 LFVSGGRVGD----VFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDA 426
+ +SGG D V+ + W + + P + G A I V + +++ E+
Sbjct 432 IAISGGTYQDWVETVYTSEWNMHTETPV-----YEGGMSAEIEFQEVVS----NSATEEE 482
Query 427 NLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDF 486
LG LA + + EP M IA + P Y QG D+ S+ DD
Sbjct 483 PLGTLAG--RGFASNKKGGQLHIKVTEPCYIMGIASITPRVDYCQGNDWDITSLDTMDDI 540
Query 487 N-PELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEEVAW 545
+ P+L+ IG+Q + + + + A +N +VG++ +W
Sbjct 541 HKPQLDSIGYQDLMQEQ----------MNAQASRN----------------LAVGKQPSW 574
Query 546 SWLRTDYPRLHGDFA-QNGNYQ-YWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQY 603
T + + +G FA ++G + + VL R F DAGT + STYI+P + Y
Sbjct 575 INYMTSFNKTYGTFANEDGEGEAFMVLNRYFDIKEIDAGTETGVKVYNTSTYIDPSQYNY 634
Query 604 VFVDQTLMAGNFAYYGYFDLKVTSSLSANYMPYL 637
+F + + NF F ++ +SA+ +P L
Sbjct 635 IFAETGTKSMNFWVQLGFGIEARRVMSASQIPNL 668
Score = 23.1 bits (48), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/56 (25%), Positives = 24/56 (43%), Gaps = 0/56 (0%)
Query 16 RAIHSRVAPSYGTIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKL 71
R+ H ++ G VP G+ +ID+ + + P +GPL +KL
Sbjct 27 RSTHDLSYAWRSSMGVGTLVPCMKLLGLPGDTFDIKIDNKVLTHPTVGPLFGSYKL 82
> Alpavirinae_Human_feces_A_032_Microviridae_AG0217_putative.VP1
Length=657
Score = 73.2 bits (178), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 79/307 (26%), Positives = 129/307 (42%), Gaps = 52/307 (17%)
Query 311 GGLLSVPYSPDLFSNIIKQGSSPAVEIEVMNALDASTDTGFS---VAVPELRLKTKLQNW 367
GGL Y DLF+N + + ++ N ++A TD S + + L L K+ N
Sbjct 367 GGLCLKTYQSDLFNNWVNKE-----WVDGDNGINAVTDVDVSEGKLNLDALNLAQKVYNM 421
Query 368 MDRLFVSGGRVGDVFRTLWGVKSSAPY---VNKPDFLGVWQASINPSNVRAMANGSASGE 424
++R+ +SGG D T++ + Y P + G +I+ V + + +ASG
Sbjct 422 LNRIAISGGSYKDWIETVY----TTDYYFRAETPVYEGGMSTTIDFEAVVSNSASTASGI 477
Query 425 DANLGQLAACVDRYCDFSGHSG-IDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFG 483
+ LG LA R + + G I EP + IA + P Y QG D+ +
Sbjct 478 EEPLGSLAG---RGFNQGKNGGKIRIKCNEPCYIIGIASITPNVDYSQGNDWDMTQLKTM 534
Query 484 DDFN-PELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEE 542
DD + P+L+GIG+Q + ++ + D ++G++
Sbjct 535 DDLHKPQLDGIGYQDLLSNQMNGLANPTD--------------------------AIGKQ 568
Query 543 VAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDWQ 602
AW TD + + DFA Y VL R + + GT + STYI+P D+
Sbjct 569 PAWLNYMTDVNKTYADFAAGETESYMVLNRIY-DVNEETGTII-----NPSTYISPKDYT 622
Query 603 YVFVDQT 609
Y+F T
Sbjct 623 YIFATNT 629
Lambda K H a alpha
0.321 0.140 0.442 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 59263415