bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_1
Length=272
Score E
Sequences producing significant alignments: (Bits) Value
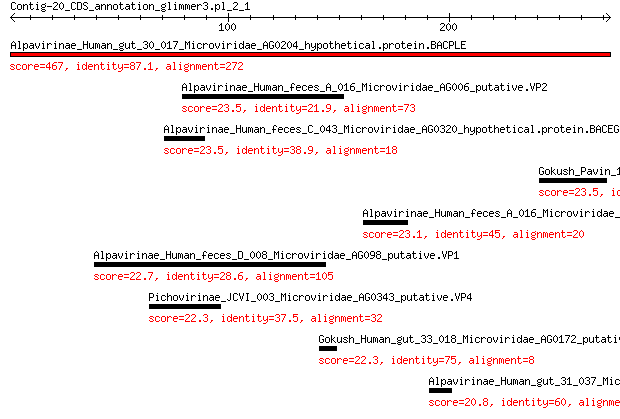
Alpavirinae_Human_gut_30_017_Microviridae_AG0204_hypothetical.p... 467 1e-166
Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2 23.5 1.9
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 23.5 2.0
Gokush_Pavin_110_Microviridae_AG052_putative.VP2 23.5 2.3
Alpavirinae_Human_feces_A_016_Microviridae_AG002_putative.VP4 23.1 2.7
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 22.7 3.9
Pichovirinae_JCVI_003_Microviridae_AG0343_putative.VP4 22.3 4.3
Gokush_Human_gut_33_018_Microviridae_AG0172_putative.VP2 22.3 4.4
Alpavirinae_Human_gut_31_037_Microviridae_AG0298_hypothetical.p... 20.8 9.2
> Alpavirinae_Human_gut_30_017_Microviridae_AG0204_hypothetical.protein.BACPLE
Length=422
Score = 467 bits (1202), Expect = 1e-166, Method: Compositional matrix adjust.
Identities = 237/272 (87%), Positives = 257/272 (94%), Gaps = 2/272 (1%)
Query 1 LKNAQQERERSAAALNDAEADWYKSQTLDKDLRERLMKAQAGLAEQGITESSSRASLNTA 60
LK+AQQERERSAA+LNDAEADWY+SQTLDKDLRERLMKAQAGLAE GITESSSRASLN A
Sbjct 152 LKDAQQERERSAASLNDAEADWYRSQTLDKDLRERLMKAQAGLAEAGITESSSRASLNAA 211
Query 61 ISLSYSIDNELKDAAIGYNLEMIKADLGKAKEEYYQLKARTGYIDDQIEAELQLLTARAL 120
I+LSYSIDNELKDAA GYNLEMIKA+LGKAKEEYYQLK RTGYIDD +E ELQLLTARA+
Sbjct 212 ITLSYSIDNELKDAAFGYNLEMIKANLGKAKEEYYQLKTRTGYIDDLLEGELQLLTARAI 271
Query 121 YLKSSSSNQEQLARVNELTADDLENWFDVNWNTKVQVPIINEKGKVERTVEMTGKEIRRE 180
YLKSS+SNQEQLARVN+LTADDLENWFDVNWNT+V+VPII+EKGK+ERTV+MTGKEIRRE
Sbjct 272 YLKSSASNQEQLARVNDLTADDLENWFDVNWNTQVEVPIIDEKGKIERTVKMTGKEIRRE 331
Query 181 YMKLNLQDFQYDMYTNRWELRSEKNRFGYSVVNTavsgaisaaghiagaKVLSTAPPVQR 240
YMKLNLQDFQYDMYTNRW LRSEKNRFGYS+VNTAV+GAISAAGH+ GAKVLSTAPPVQR
Sbjct 332 YMKLNLQDFQYDMYTNRWSLRSEKNRFGYSIVNTAVNGAISAAGHVVGAKVLSTAPPVQR 391
Query 241 FDDVTEEFVPTSSSLGGGWTKHTSTTSRQIRR 272
FDDVTE+ VP+ G GWTKHT+TTSRQ RR
Sbjct 392 FDDVTEDLVPSPD--GAGWTKHTTTTSRQFRR 421
> Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2
Length=367
Score = 23.5 bits (49), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/73 (22%), Positives = 32/73 (44%), Gaps = 0/73 (0%)
Query 79 NLEMIKADLGKAKEEYYQLKARTGYIDDQIEAELQLLTARALYLKSSSSNQEQLARVNEL 138
N+ KA+ K EE+ Q T Y+D + +Q++T + ++ +E + +
Sbjct 209 NIIEAKANAKKTVEEFKQAMLNTEYLDKTQQQRIQMVTDQLSLIQKQGLKEEAVIDLTNA 268
Query 139 TADDLENWFDVNW 151
A + D+ W
Sbjct 269 QASKVRKEIDILW 281
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 23.5 bits (49), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 7/18 (39%), Positives = 14/18 (78%), Gaps = 0/18 (0%)
Query 71 LKDAAIGYNLEMIKADLG 88
LK+ +GY +++++ DLG
Sbjct 309 LKNKGLGYEIDLLEGDLG 326
> Gokush_Pavin_110_Microviridae_AG052_putative.VP2
Length=257
Score = 23.5 bits (49), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 12/31 (39%), Positives = 20/31 (65%), Gaps = 1/31 (3%)
Query 241 FDDVTEEFVPTSSSLGGGWTKH-TSTTSRQI 270
FD +T E ++++GG + T+TT+RQI
Sbjct 2 FDWLTPEIASAAATVGGFLGQQETNTTNRQI 32
> Alpavirinae_Human_feces_A_016_Microviridae_AG002_putative.VP4
Length=355
Score = 23.1 bits (48), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 161 NEKGKVERTVEMTGKEIRRE 180
N+K K +R E+ GKE+R++
Sbjct 309 NKKSKAQRLFELYGKELRKK 328
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 22.7 bits (47), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 60/119 (50%), Gaps = 16/119 (13%)
Query 39 AQAGLAEQGITESSSRASLN----TAISLS--YSIDNELKDAAIGYNLEMIKADLGKAKE 92
++AGL T ++ +SL+ +A+S S Y + + + A +G + +++ + +A +
Sbjct 307 SRAGLVAVSGTSPAAGSSLDMQTTSALSASTKYGVYAQ-QVAGLGSSFSILQLRMAEAVQ 365
Query 93 EYYQLK------ARTGYIDDQIEAEL-QLLTARALYLKSSSSNQEQLARVN-ELTADDL 143
+Y ++ AR G I L +L+ + +YL SSSN + VN +T D++
Sbjct 366 KYREVSQFADQDAR-GQIMAHFGVSLSPVLSDKCMYLGGSSSNIDLSEVVNTNITGDNI 423
> Pichovirinae_JCVI_003_Microviridae_AG0343_putative.VP4
Length=275
Score = 22.3 bits (46), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 12/32 (38%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 64 SYSIDNELKDAAIGYNLEMIKADLGKAKEEYY 95
++ I E+KDA + + M AD K EYY
Sbjct 28 TFRILEEMKDAESAHFVTMTYADHELPKIEYY 59
> Gokush_Human_gut_33_018_Microviridae_AG0172_putative.VP2
Length=242
Score = 22.3 bits (46), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 6/8 (75%), Positives = 8/8 (100%), Gaps = 0/8 (0%)
Query 141 DDLENWFD 148
D++ENWFD
Sbjct 207 DEIENWFD 214
> Alpavirinae_Human_gut_31_037_Microviridae_AG0298_hypothetical.protein
Length=115
Score = 20.8 bits (42), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 6/10 (60%), Positives = 9/10 (90%), Gaps = 0/10 (0%)
Query 191 YDMYTNRWEL 200
YD+ T+RWE+
Sbjct 59 YDIRTDRWEI 68
Lambda K H a alpha
0.311 0.126 0.347 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 22327780