bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
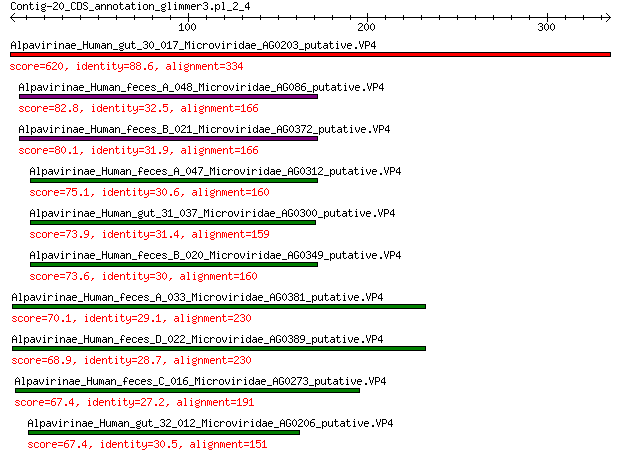
Query= Contig-20_CDS_annotation_glimmer3.pl_2_4
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4 620 0.0
Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4 82.8 9e-20
Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4 80.1 7e-19
Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4 75.1 4e-17
Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4 73.9 7e-17
Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4 73.6 2e-16
Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4 70.1 2e-15
Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4 68.9 5e-15
Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4 67.4 2e-14
Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4 67.4 2e-14
> Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4
Length=363
Score = 620 bits (1598), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 296/336 (88%), Positives = 312/336 (93%), Gaps = 2/336 (1%)
Query 1 MNRPWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYY 60
MNRPWDYFTQRIMVPCG CEECLRQQRN+WY+RLERETKY KSLH+NSVFVTITIAPEYY
Sbjct 28 MNRPWDYFTQRIMVPCGHCEECLRQQRNDWYIRLERETKYHKSLHNNSVFVTITIAPEYY 87
Query 61 DSALLNPSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYN 120
DSAL +PS+FIRMWFER+RRRFGHSIKHAVFQEFGMHPEQG EPRLHFHG+LWDVS SYN
Sbjct 88 DSALQDPSSFIRMWFERIRRRFGHSIKHAVFQEFGMHPEQGREPRLHFHGILWDVSYSYN 147
Query 121 AIREAVKDLGFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVGKLKTNLYDF 180
AIREAVKDLGFVWI+S+TDKRLRYVVKYVGKSVYMDE SA FAKSLPIT GKL TNLYDF
Sbjct 148 AIREAVKDLGFVWIASVTDKRLRYVVKYVGKSVYMDESSAAFAKSLPITAGKLNTNLYDF 207
Query 181 LQNSRYRRKFISAGVGDYLGDFKAPGVASGLWSYTDHKTGCVYRYRIPRYYDKYLSQDAL 240
LQN +Y RKF+S GVGDYLGDFKAPGV SGLWSYTD+KTG VYRYRIPRYYDKYLSQDAL
Sbjct 208 LQNRKYHRKFVSPGVGDYLGDFKAPGVTSGLWSYTDYKTGVVYRYRIPRYYDKYLSQDAL 267
Query 241 LFRKISTAWTYASAFGGSMALGFLREVAERVLRPSDFSRVVKGGFSRLVKLREFLSKVKA 300
FRKISTAWTYASAFG S+ALGFLREVA RVL PS FSRVVKGGFSRLVKLR+FLSKVK
Sbjct 268 FFRKISTAWTYASAFGSSLALGFLREVAARVLSPSSFSRVVKGGFSRLVKLRDFLSKVKV 327
Query 301 RPS--FLAVTSDVIDFWVDCFGIDSSNPFFNKIVYG 334
+ S F+AVT DVIDFWVDCFGIDSSNPFFNKIVYG
Sbjct 328 KVSQRFIAVTPDVIDFWVDCFGIDSSNPFFNKIVYG 363
> Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4
Length=332
Score = 82.8 bits (203), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 54/177 (31%), Positives = 88/177 (50%), Gaps = 25/177 (14%)
Query 6 DYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALL 65
D T I CG+C EC +Q++ EW VR+ E + + N+ F+T+TI+ E Y+ L
Sbjct 28 DPRTAYITAACGKCLECRKQKQREWLVRMSEELRTEP----NAYFMTLTISDENYE-ILK 82
Query 66 N----------PSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDV 115
N + IR+ ER+R++ G SIKH E G + RLH HG++W +
Sbjct 83 NICKSEDDNTIATKAIRLMLERIRKKTGKSIKHWFITELGHEKTE----RLHLHGIVWGI 138
Query 116 SCSYNAIREAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
+ + E + G + + + +K + YV KY+ K +DE+ DF + + G
Sbjct 139 GT--DQLIEEKWNYGITYTGNYVNEKTINYVTKYMTK---IDEKHPDFVGKVLCSRG 190
> Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4
Length=332
Score = 80.1 bits (196), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 53/177 (30%), Positives = 86/177 (49%), Gaps = 25/177 (14%)
Query 6 DYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYY----- 60
D T I CG+C EC +Q++ EW VR+ E + + N+ F+T+TI+ E Y
Sbjct 28 DPRTAYITAACGKCLECRKQKQREWLVRMSEELRTEP----NAYFMTLTISDENYEILKN 83
Query 61 -----DSALLNPSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDV 115
D + A IR+ ER+R++ G SI+H E G + RLH HG++W +
Sbjct 84 ICKSEDENTIATKA-IRLMLERIRKKIGKSIRHWFITELGHEKTE----RLHLHGIVWGI 138
Query 116 SCSYNAIREAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
IRE + G + + + +K + Y+ KY+ K +DE +F + + G
Sbjct 139 GTD-QLIREKW-NYGITYTGNFVNEKTINYITKYMTK---IDEEHPNFVGKVLCSKG 190
> Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4
Length=332
Score = 75.1 bits (183), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 49/171 (29%), Positives = 85/171 (50%), Gaps = 25/171 (15%)
Query 12 IMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLN----- 66
I CG+C EC +Q++ +W VR+ E + + N+ F+T+TI + Y S L N
Sbjct 34 ITAACGKCMECRQQKQRQWLVRMSEELRQEP----NAYFITLTIDDKSY-SELSNTYNIT 88
Query 67 -----PSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYNA 121
+ IR+ ER+R++ G SIKH E G + RLH HG++W + +
Sbjct 89 DNNEIATKAIRLCLERIRKQTGKSIKHWFITELGHEKTE----RLHLHGIVWGIGT--DK 142
Query 122 IREAVKDLGFVWIS-SITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
+ + + G + + +K ++Y+ KY+ K +DE+ DF + + G
Sbjct 143 LITSKWNYGITFTGFFVNEKTIQYITKYMTK---IDEQHKDFIGKVLCSKG 190
> Alpavirinae_Human_gut_31_037_Microviridae_AG0300_putative.VP4
Length=304
Score = 73.9 bits (180), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 50/169 (30%), Positives = 87/169 (51%), Gaps = 26/169 (15%)
Query 12 IMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDS------ALL 65
+ CG+C EC +Q+ W VRL E + N++FVT+TI+ E ++ L
Sbjct 34 VTAACGKCYECRKQKGRAWQVRLSEEVRSD----PNAIFVTLTISDESWEKIKNTYIQLS 89
Query 66 NPSA---FIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYNAI 122
+ +R++ ERVR++ S+KH + E G N R H HG++W + A+
Sbjct 90 DEDCIKKMVRLFLERVRKKTKKSLKHWLTTERG----GTNTERYHLHGLIWGENTE--AL 143
Query 123 REAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDERSADFAKSLPITV 170
+++ GFV+I + + + + Y+ KY+ K+ D++ DF PIT+
Sbjct 144 TKSLWQYGFVFIGTFVNECTVNYITKYITKT---DKKHKDFE---PITL 186
> Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4
Length=332
Score = 73.6 bits (179), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 48/170 (28%), Positives = 84/170 (49%), Gaps = 23/170 (14%)
Query 12 IMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYD--SALLN--- 66
+ CG+C EC +Q++ +W VR+ E + + N+ F+T+TI E Y+ + + N
Sbjct 34 VTAACGKCMECRQQKQRQWLVRMSEELRQ----NPNAYFMTLTIDDENYNKLANICNSKD 89
Query 67 ----PSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYNAI 122
+ +R+ ER+R++ G SIKH E G + RLH HG++W + I
Sbjct 90 NNEIATKAVRLMLERIRKKTGKSIKHWFITELGHEKTE----RLHLHGIVWGIGTD-QLI 144
Query 123 REAVKDLGFVWISS-ITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
E + GFV+ + + + + Y+ KY+ K +D DF + + G
Sbjct 145 SEKW-NYGFVYTGNFVNEATINYITKYMTK---VDIDHPDFVGQVLCSKG 190
> Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4
Length=307
Score = 70.1 bits (170), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 67/251 (27%), Positives = 106/251 (42%), Gaps = 41/251 (16%)
Query 2 NRPWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYD 61
N+P DY ++ VPCG+C +C+++++ W+VR K N+ F T T+ PE+Y+
Sbjct 29 NQP-DY---KLQVPCGKCVQCIKKRQQHWFVRAHNIYKRLGYNLSNTYFCTFTLKPEFYE 84
Query 62 SALLNPSAFIRMWFERVRR------------RFGH---SIKHAVFQEF--GMHPEQ---G 101
+ P AFIR + +R+R+ RF + S + E G Q
Sbjct 85 AFCKEPYAFIRRFIDRMRKDQSLRYRDPDTGRFRYRKLSFPYLFVLEVADGKRAAQRKLS 144
Query 102 NEPRLHFHGVLWDVSCSYNAIREAVKDLGFVWISSITD-KRLRYVVKYVGKSVYMDERSA 160
+E RLH H +++ + ++R G W+S + +RY +KYV K +SA
Sbjct 145 SEHRLHIHAIMFGCPLPWWSVRHYWMSFGLAWVSRLRHFGGVRYAMKYVTK------KSA 198
Query 161 DFAKSLPITVGKLKTNLYDFLQNSRYRRKFISAGVGDYLGDFKAPGVASGLWSYTDHKTG 220
+P + L LY R +S D L + G D+
Sbjct 199 VHRNDVPNDILDLHGRLYVSHGFGR-----LSESERDALRAYMMTGCKQWFSILIDNHP- 252
Query 221 CVYRYRIPRYY 231
Y IPRYY
Sbjct 253 ----YSIPRYY 259
> Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4
Length=307
Score = 68.9 bits (167), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 66/253 (26%), Positives = 106/253 (42%), Gaps = 45/253 (18%)
Query 2 NRPWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYD 61
N+P DY ++ VPCG+C +C+++++ W+VR K N+ F T T+ PE+Y+
Sbjct 29 NQP-DY---KLQVPCGKCVQCIKKRQQHWFVRAHNIYKRLGYNLSNTYFCTFTLKPEFYE 84
Query 62 SALLNPSAFIRMWFERVRR------------RFGH-SIKHAVFQEFGMHPEQG------- 101
+ P AFIR + +R+R+ RF + I F F + G
Sbjct 85 AFCKEPYAFIRRFIDRMRKDQLLRYRNPDTGRFCYRKISFPYF--FVLEVADGKRAAQRR 142
Query 102 --NEPRLHFHGVLWDVSCSYNAIREAVKDLGFVWISSITD-KRLRYVVKYVGKSVYMDER 158
+E RLH H +++ + +R G W++ + +RY +KY+ K +
Sbjct 143 LHSEHRLHLHAIMFGCPLPWWRVRHYWMSFGLAWVNPLRHFGAVRYAMKYITK------K 196
Query 159 SADFAKSLPITVGKLKTNLYDFLQNSRYRRKFISAGVGDYLGDFKAPGVASGLWSYTDHK 218
SA +P V L LY R +S D L + G D+
Sbjct 197 SAVHWNDVPKEVLDLHGRLYVSHGFGR-----LSESEKDTLRAYMMTGCKQWFSILIDN- 250
Query 219 TGCVYRYRIPRYY 231
+ Y IPRYY
Sbjct 251 ----HPYSIPRYY 259
> Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4
Length=304
Score = 67.4 bits (163), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/204 (25%), Positives = 91/204 (45%), Gaps = 23/204 (11%)
Query 4 PWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSA 63
P DY + VPCG C C + N++ +RL E + K +FVT+T +
Sbjct 19 PPDY---NLEVPCGYCHSCQKSYNNQYRIRLLYELR--KYPSGTCLFVTLTFDDDNLKKF 73
Query 64 LLNPSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCS----- 118
+ + +R++ +R+R+ +G I+H EFG R H+HG+L++V +
Sbjct 74 SKDTNKAVRLFLDRLRKDYGKQIRHWFVCEFGTL-----RCRPHYHGILFNVPQALIDGY 128
Query 119 ------YNAIREAVKDLGFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVGK 172
++ + + GFV++ ++++ Y+ KYV KS+ D+ S I
Sbjct 129 SPDVPGHHPLLASRWKYGFVFVGYVSEETCSYITKYVTKSINGDKVRPRVISSFGIGSNY 188
Query 173 LKTNLYDF--LQNSRYRRKFISAG 194
L T L N RY+ + G
Sbjct 189 LDTEESSLHKLGNQRYQPFMVLNG 212
> Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4
Length=306
Score = 67.4 bits (163), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 46/179 (26%), Positives = 83/179 (46%), Gaps = 35/179 (20%)
Query 11 RIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLNPSAF 70
++ VPCG C+ECL++++ +W++R + K K +F T +I PE Y+ P
Sbjct 34 KLRVPCGHCDECLKKRQQQWFLRADHVVKRLKLRPDQCLFCTFSIKPEVYEQTKERPYLA 93
Query 71 IRMWFERVRRRFGHSIKHAVFQEFGMHP------------------------EQGNEP-- 104
IR + +R+R KH F+E G E+G E
Sbjct 94 IRRFMDRLR-------KHPRFREKGASGRYRYRKVRFPYIFVVEFADGKRARERGLESTH 146
Query 105 RLHFHGVLWDVSCSYNAIREAVKD-LGFVWISSI-TDKRLRYVVKYVGKSVYMDERSAD 161
R+HFH +L++ + +R+ + +G W+ + ++ +RYV+KY+ K + +D
Sbjct 147 RMHFHAILFNCPLYWWQVRDLWESCVGRAWVDPLHSEAGIRYVLKYMTKDCKAHQYISD 205
Lambda K H a alpha
0.326 0.140 0.444 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 28557990