bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_5
Length=648
Score E
Sequences producing significant alignments: (Bits) Value
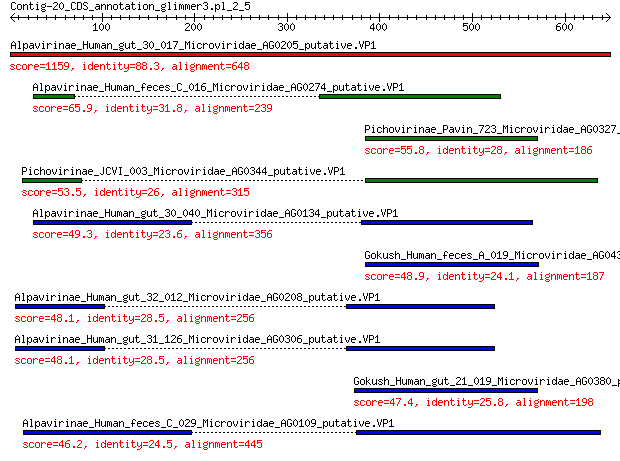
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 1159 0.0
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 65.9 5e-13
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 55.8 6e-10
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 53.5 3e-09
Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1 49.3 8e-08
Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1 48.9 8e-08
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 48.1 2e-07
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 48.1 2e-07
Gokush_Human_gut_21_019_Microviridae_AG0380_putative.VP1 47.4 2e-07
Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1 46.2 5e-07
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 1159 bits (2998), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 572/648 (88%), Positives = 609/648 (94%), Gaps = 1/648 (0%)
Query 1 MGKQPFISHAVNGYSRYDMPENKAFSVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLG 60
MGKQPFISH+VNGYSRYDMPENKAFS+TPGIIYPVRIQFVNARDRVTLHQG+DVRSNPLG
Sbjct 1 MGKQPFISHSVNGYSRYDMPENKAFSITPGIIYPVRIQFVNARDRVTLHQGVDVRSNPLG 60
Query 61 VPSFNPYVLRLHRFWVPLQLYHPEMRVNSSKFDMNDLSFNFIPCCVDNKGLDAYTSFMYP 120
VPSFNPYVLRLHRFWVPLQLYHPEMRVNSSKFDMN+L++NFIP VDNKG YTS+MYP
Sbjct 61 VPSFNPYVLRLHRFWVPLQLYHPEMRVNSSKFDMNNLTYNFIPGVVDNKGGGNYTSYMYP 120
Query 121 RPGVAAFFSQVMPFNHRAALPNSLMSWLRVANSPIINYPVNTVPSLSSFLKTTSKFVTVN 180
R G AAFFSQVMPFN RAALPNSLMSWLR+ANSPI NY N +PS KT+ KF++VN
Sbjct 121 RSGTAAFFSQVMPFNCRAALPNSLMSWLRIANSPIFNYSGNPLPSGGVLFKTSPKFLSVN 180
Query 181 ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPTFYTTSTSSVSKPEYRSQASYFWQRYG 240
ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRP FYT ++SSV+ EYRSQASYFWQRYG
Sbjct 181 ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPVFYTGTSSSVTNVEYRSQASYFWQRYG 240
Query 241 NLEFLDHYFETMFYPRDRNVDSNRDELSWNRSDLFIEILRSDLFNTGTIAAAPDFNKLVQ 300
NLEFLDHYFETMFYPRDR V ++RDELSWNRSDLF+EILRSDLFN T AA+PDF K+ Q
Sbjct 241 NLEFLDHYFETMFYPRDRIVAADRDELSWNRSDLFVEILRSDLFNKDTTAASPDFTKMPQ 300
Query 301 TFPQNVNFNVPAYPYDVQEPKVNWNNGSGTDTGTPSKVYFAATLNVPFLAAHPMAVCPSS 360
+P+ +NFNVPAYPYDVQEPKV+WNNG+GTD PSKV+FAATLNVPFLAAHPMAVCPSS
Sbjct 301 MYPEAMNFNVPAYPYDVQEPKVDWNNGTGTDVNVPSKVFFAATLNVPFLAAHPMAVCPSS 360
Query 361 PDRFSRLMPPGDSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI 420
PDRFSRLMPPGDSNSDVDFTG+KTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI
Sbjct 361 PDRFSRLMPPGDSNSDVDFTGLKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI 420
Query 421 EHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFK 480
EHVDRPKLLFSSSVMVNSQVVMNQAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFK
Sbjct 421 EHVDRPKLLFSSSVMVNSQVVMNQAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFK 480
Query 481 EPGYIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASES 540
EPGYIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVP WR+G+GWK+ + S
Sbjct 481 EPGYIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGFGWKSDAVS 540
Query 541 QSMTVAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYMIGLSSNPNEIS 600
S++VAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFY+IGLSS+ NEIS
Sbjct 541 -SVSVAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYLIGLSSDLNEIS 599
Query 601 PSMLFTNLSTVNNPFASDMEDNFFVNMSYKVVVKNLVNKSFATRLSSR 648
PSMLFTNL+TVNNPF+SDMEDNFFVNMSYKVVVKNLVNKSFATRLSSR
Sbjct 600 PSMLFTNLATVNNPFSSDMEDNFFVNMSYKVVVKNLVNKSFATRLSSR 647
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 65.9 bits (159), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 65/216 (30%), Positives = 92/216 (43%), Gaps = 28/216 (13%)
Query 335 PSKVYFAATLNVPFLAAHPMAVCPSSPDRFSRLMPPG-------------DSNSDVDFTG 381
P+ F A + P SPD F ++ G DSN+D F+
Sbjct 296 PTSNSFVAMACAGIQGYGGLLSVPYSPDLFGNIIKQGSSPTAEIEVIKALDSNTDTGFS- 354
Query 382 VKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIE--HVDRPKLLFsssvmvnsq 439
+P+L + T++Q + D + SG R D T + +K +V++P L +N
Sbjct 355 -IAVPELRLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASINPS 413
Query 440 vvmnqAGQSGFAGGEAAALGQMGGSIA----FNTVLGREQTYYFKEPG--YIFDMLTIRP 493
V A +G A GE A LGQ+ + F+ G + YY KEPG + ML P
Sbjct 414 NVRAMA--NGSASGEDANLGQLAACVDRYCDFSDHSGID--YYAKEPGTFMLITMLVPEP 469
Query 494 VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWR 529
Y G+ PD D FNP N IG+Q VP R
Sbjct 470 AYS-QGLHPDLASISFGDDFNPELNGIGFQLVPRHR 504
Score = 23.1 bits (48), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 11/44 (25%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 26 SVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFNPYVL 69
++ PG+ PV + +N +R+ ++S P+ P N + L
Sbjct 28 TIHPGLAIPVHHRHLNVGERIRGRIDELLQSQPMLGPLMNGFKL 71
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 55.8 bits (133), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 52/189 (28%), Positives = 85/189 (45%), Gaps = 13/189 (7%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLLFsssvmvnsqvv 441
TI L A +LQE+ + G+RY + + T F S + + RP+ + V V
Sbjct 268 TINDLRRAFKLQEWLEKNARGGTRYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEV 327
Query 442 mnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP-VYFWTGI 500
+N GQ G G M G + G+ +YY +E GYI ++++ P + GI
Sbjct 328 LNTTGQDG-----GLPQGNMAGH-GISVTSGKSGSYYCEEHGYIIGIMSVMPKTAYQQGI 381
Query 501 RPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPCYNEFRSSYDE 560
+L+ DYF P + +IG Q+V + Y + A + T P Y E++
Sbjct 382 PRTFLKTDSLDYFWPTFANIGEQEVAKQEL-YAYTANAND---TFGYVPRYAEYKYMPSR 437
Query 561 VLGSLQSTL 569
V G +++L
Sbjct 438 VAGEFRTSL 446
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 53.5 bits (127), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 66/256 (26%), Positives = 110/256 (43%), Gaps = 31/256 (12%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFF--ASKIEHVDRPKLLFsssvmvnsqvv 441
TI L A +LQE+ +L+ GSRY + + TFF S + + RP+ + + V +
Sbjct 265 TITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISEI 324
Query 442 mnqAGQS-GFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP-VYFWTG 499
+N G++ G G + G G+ G YY +E G+I +L++ P + G
Sbjct 325 LNTTGETAGLPQGNMSGHGVAVGT-------GNVGKYYCEEHGFIIGILSVTPNTAYQQG 377
Query 500 IRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPCYNEFRSSYD 559
I YL D++ P + IG Q + I Y + + T P Y E++
Sbjct 378 IPKHYLRTDKYDFYWPQFAHIGEQPIVNNEI-YAYDPTGDE---TFGYTPRYAEYKYMPS 433
Query 560 EVLGSLQSTLTPKASVPLQSYWVQQRDFYMIGLSSNPNEISPSMLFTNLSTVNNPFA-SD 618
V G +S+L W R F SS P +S + + + FA +D
Sbjct 434 RVAGEFRSSL---------DEWHAARIF-----SSLPT-LSQDFIEVDADDFDRIFAVTD 478
Query 619 MEDNFFVNMSYKVVVK 634
+DN ++++ KV +
Sbjct 479 GDDNLYMHVLNKVKAR 494
Score = 26.2 bits (56), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 14 YSRYDMPENKAFSVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFNPYVLRLHR 73
Y+ +D+ + S G + P+ D +TL V+ PL P + + + +H
Sbjct 17 YNTFDLTHDVKMSGRMGELLPIMNLECIPGDNITLGADSMVKFAPLLAPVMHRFNVTMHY 76
Query 74 FWVP 77
F+VP
Sbjct 77 FFVP 80
> Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1
Length=679
Score = 49.3 bits (116), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 50/192 (26%), Positives = 88/192 (46%), Gaps = 20/192 (10%)
Query 380 TGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASK-IEHVDRPKLLFsssvmvns 438
+G T+ L +A ++ + I S Y+ W+ T + S + H++ P L SS+ +
Sbjct 434 SGSFTLDTLNLAKKVYTMLNRIAVSDGSYNAWIQTVYTSGGLNHIETPLYLGGSSLEIEF 493
Query 439 qvvmnqAGQS----GFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP- 493
Q V+N +G G G A GG+I F EPGYIF + +I P
Sbjct 494 QEVINNSGTEDQPLGALAGRGVATNHKGGNIVFKA----------DEPGYIFCITSITPR 543
Query 494 VYFWTGIRPD-YLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPCYN 552
V ++ G D YLE D P + IG+QD + + E ++++ K+P +
Sbjct 544 VDYFQGNEWDLYLESL-DDLHKPQLDGIGFQDRLYRHLNS--SCVREDLNISIGKQPAWV 600
Query 553 EFRSSYDEVLGS 564
++ ++ ++ G+
Sbjct 601 QYMTNVNKTYGN 612
Score = 29.3 bits (64), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 34/171 (20%), Positives = 62/171 (36%), Gaps = 27/171 (16%)
Query 26 SVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFNPYVLRLHRFWVPLQLYHPEM 85
++ PG++ P + + D + ++P P F + + F+ P++LY+ +
Sbjct 37 TMAPGVLVPTMTELLLPGDTYPIQIRCHTLTHPTIGPLFGSFKQQNDFFFCPIRLYNSML 96
Query 86 RVNSSKFDMNDLSFNFIPCCVDNKGLDAYTSFMYPRPGVAAFFSQVMPFNHRAALPNSLM 145
N+ ++ F +D D + + R P+SL+
Sbjct 97 HNNALNIGLDMKKVKFPVIKIDASDWDLNKKMVGSNNTLL-----------RQIHPSSLL 145
Query 146 SWLRVANSPIINYPVNTVPSLSSFLKTTSKFVTVNADTYLGYWDIVRNYYS 196
S+L SF K T T N L Y+DI +NYY+
Sbjct 146 SYL----------------GYKSFNKETGVDWTWNITKALMYYDIFKNYYA 180
> Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1
Length=569
Score = 48.9 bits (115), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 45/192 (23%), Positives = 92/192 (48%), Gaps = 16/192 (8%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI--EHVDRPKLLFsssvmvnsqvv 441
TI Q+ A ++Q+Y + + GSRY + +Y+ F ++I + V P+ L +M+N V
Sbjct 318 TINQIRQAFQVQKYFEELARGGSRYREQIYSLFRTRISDKTVQIPEYLGGDRIMINMSQV 377
Query 442 mnqAGQSGFA-GGEAAALGQMG-GSIAFNTVLGREQTYYFKEPGYIFDMLTIRPVY-FWT 498
+ +G + + G +A+ G G AF T F+E G+I + +R + +
Sbjct 378 VQTSGTTDVSPQGNVSAMSVTGFGKSAF--------TKSFEEHGFIIGVCCVRHDHTYQQ 429
Query 499 GIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPCYNEFRSSY 558
G+ + DY+ P++ ++G Q V + + +E+ + + C+ ++R
Sbjct 430 GLERMFSRKNKLDYYFPVFANLGEQAVLKKEL---YAQGTETDNEAFGYQECWADYRMKP 486
Query 559 DEVLGSLQSTLT 570
+ + G+ +S T
Sbjct 487 NRICGAFRSNAT 498
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 48.1 bits (113), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/177 (29%), Positives = 76/177 (43%), Gaps = 22/177 (12%)
Query 364 FSRLMPP------------GDSNSDVDFTG-VKTIP--QLAVATRLQEYKDLIGASGSRY 408
F R +PP DS VD K+I +A + +Q + DL A GSRY
Sbjct 261 FQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRY 320
Query 409 SDWLYTFF-ASKIEHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAF 467
SD++ + F S+++H P L S + S V+ G A A GQ G F
Sbjct 321 SDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETF 380
Query 468 NTVLGREQTYYFKEPGYIFDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQ 523
R ++Y+F E GY M ++ P V + G+ P E D + P ++I +
Sbjct 381 -----RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGDVYVPALDNIAME 432
Score = 35.0 bits (79), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/96 (23%), Positives = 45/96 (47%), Gaps = 0/96 (0%)
Query 6 FISHAVNGYSRYDMPENKAFSVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFN 65
F++ + N S ++ ++ PG + P+ + A D + V++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYVLRLHRFWVPLQLYHPEMRVNSSKFDMNDLSFNF 101
+ L L F+VP +LY+ + ++++ N S F
Sbjct 65 GFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKF 100
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 48.1 bits (113), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/177 (29%), Positives = 76/177 (43%), Gaps = 22/177 (12%)
Query 364 FSRLMPP------------GDSNSDVDFTG-VKTIP--QLAVATRLQEYKDLIGASGSRY 408
F R +PP DS VD K+I +A + +Q + DL A GSRY
Sbjct 261 FQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRY 320
Query 409 SDWLYTFF-ASKIEHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAF 467
SD++ + F S+++H P L S + S V+ G A A GQ G F
Sbjct 321 SDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETF 380
Query 468 NTVLGREQTYYFKEPGYIFDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQ 523
R ++Y+F E GY M ++ P V + G+ P E D + P ++I +
Sbjct 381 -----RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGDVYVPALDNIAME 432
Score = 35.0 bits (79), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/96 (23%), Positives = 45/96 (47%), Gaps = 0/96 (0%)
Query 6 FISHAVNGYSRYDMPENKAFSVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFN 65
F++ + N S ++ ++ PG + P+ + A D + V++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYVLRLHRFWVPLQLYHPEMRVNSSKFDMNDLSFNF 101
+ L L F+VP +LY+ + ++++ N S F
Sbjct 65 GFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKF 100
> Gokush_Human_gut_21_019_Microviridae_AG0380_putative.VP1
Length=563
Score = 47.4 bits (111), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/202 (25%), Positives = 89/202 (44%), Gaps = 14/202 (7%)
Query 372 DSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLL 429
DS D+D + + TI L A ++Q++ + + GSRY++ L +FF+ S + RP+ L
Sbjct 303 DSYVDLDTSSIFTINSLRTAFQMQKFYERLARGGSRYTEVLRSFFSVVSPDARLQRPEFL 362
Query 430 Fsssvmvnsq-vvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDM 488
S + MVN + A S G +A G T F E GY+F
Sbjct 363 GSFTKMVNVNPIAQTSATDSTSPQGNLSAYGVTAAKF-------HGFTKSFVEHGYVFGF 415
Query 489 LTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAK 547
+ R + + GI +L D++ P + +G Q + I + SE+ +
Sbjct 416 VCARADLTYQQGINKMWLRSTVYDFYWPTFAHLGEQAIELREI---YAQGSEADTTVFGY 472
Query 548 EPCYNEFRSSYDEVLGSLQSTL 569
+ Y E+R ++ G +S++
Sbjct 473 QERYAEYRYKPSQITGKFRSSV 494
> Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1
Length=640
Score = 46.2 bits (108), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 44/182 (24%), Positives = 74/182 (41%), Gaps = 29/182 (16%)
Query 15 SRYDMPENKAFSVTPGIIYPVRIQFVNARDRVTLHQGIDVRSNPLGVPSFNPYVLRLHRF 74
S +D+ + PG + P V D + + +V ++P P F + L H +
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHVY 85
Query 75 WVPLQLYHPEMRVNSSKFDMNDLSFNFIPCCVDNKGLDAYTSFMYPRPGVAAFFSQVMPF 134
VP +LY+ + N +K + D+S IP N GL +
Sbjct 86 TVPFRLYNSWLHNNRTKIGL-DMSQVKIPQL--NVGLKNTDN------------------ 124
Query 135 NHRAALPNSLMSWLRVANSPIINYPVNTVPSLSSFLKTTSKFVTVNADTYLGYWDIVRNY 194
P S W ++ S ++ Y + S + T+K+V NA +GY+DI +NY
Sbjct 125 ------PTSKNEWTQINPSCLLAYL--GIRGYGSLIGVTTKYVEKNAVPLIGYYDIFKNY 176
Query 195 YS 196
Y+
Sbjct 177 YA 178
Score = 39.7 bits (91), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 65/275 (24%), Positives = 109/275 (40%), Gaps = 24/275 (9%)
Query 375 SDVDFT-GVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIEHVDRPKLLFsss 433
S VD + G ++ L +A ++ + I SG Y DWL T F E+++R +
Sbjct 370 SSVDVSDGNLSMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGG-EYMERCETPVFEG 428
Query 434 vmvnsqvvmnqAGQSGFAGGEAAALGQMGG-SIAFNTVLGREQTYYFKEPGYIFDMLTIR 492
V S E LG + G I N G EPGYI + +I
Sbjct 429 GTSQEIVFQEVISNSAT---EQEPLGTLAGRGITTNKQRGGHIKIKVTEPGYIMCICSIT 485
Query 493 P-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDV----PFWRIGYGWKAASESQSMTVAK 547
P + + G + D D P + IGYQD W GY + A + + K
Sbjct 486 PRIDYSQGNQWDTELKTMDDLHKPALDGIGYQDSINSERAWWAGY-YNADPDKKETAAGK 544
Query 548 EPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYM-IGLSSNPNEISPSMLFT 606
+ + ++ + G+ A +++ V R++ + I S P +I+ L T
Sbjct 545 TVAWINYMTNVNRTYGNF-------AIKDNEAFMVMNRNYELEITGGSTPTKITIGDLST 597
Query 607 NL--STVNNPFA-SDMED-NFFVNMSYKVVVKNLV 637
+ + N FA + +E NF+V + + + L+
Sbjct 598 YIDPAKFNYIFADTSLEAMNFWVQTKFDIKARRLI 632
Lambda K H a alpha
0.320 0.136 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 60207434