bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
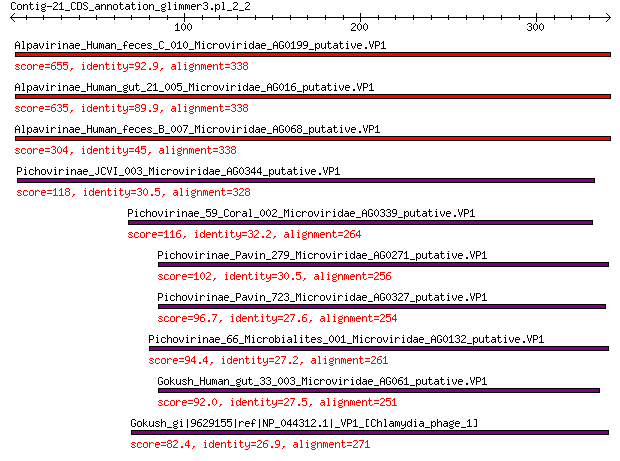
Query= Contig-21_CDS_annotation_glimmer3.pl_2_2
Length=341
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 655 0.0
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 635 0.0
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 304 1e-97
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 118 1e-31
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 116 8e-31
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 102 9e-26
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 96.7 6e-24
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 94.4 4e-23
Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1 92.0 2e-22
Gokush_gi|9629155|ref|NP_044312.1|_VP1_[Chlamydia_phage_1] 82.4 4e-19
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 655 bits (1690), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 314/338 (93%), Positives = 323/338 (96%), Gaps = 0/338 (0%)
Query 4 FVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 63
VGL +GDVVTR++DGTYS+QKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP
Sbjct 394 LVGLVMGDVVTRSEDGTYSVQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 453
Query 64 VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 123
VTAI SYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF
Sbjct 454 VTAIGSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 513
Query 124 DELLMPEFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVF 183
DELLMPEFIGGISRELSMRTVEQT+DQQ S+GQYAEALGSKTGIAGVYGSTSNNIEVF
Sbjct 514 DELLMPEFIGGISRELSMRTVEQTVDQQSGSSRGQYAEALGSKTGIAGVYGSTSNNIEVF 573
Query 184 CDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVG 243
CDEESYIIGLLTVTPVP+YTQLLPKDF YNGLLDHYQPEFDRIGFQPITYKEICPMNIV
Sbjct 574 CDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNIVA 633
Query 244 DDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSD 303
D QL+KTFGYQRPWYEYVAKYDSAHGLFR +MKNFIMSRVF GLPQLGQQFLLVD D
Sbjct 634 GDSANQLNKTFGYQRPWYEYVAKYDSAHGLFRTNMKNFIMSRVFSGLPQLGQQFLLVDPD 693
Query 304 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 341
TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 694 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 635 bits (1639), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 304/338 (90%), Positives = 319/338 (94%), Gaps = 1/338 (0%)
Query 4 FVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 63
VGLTVGDVVTR++DGT S+QKQTVLVDEDGSKYG+SYKVSEDGERLVGVDYDPVSEKTP
Sbjct 406 LVGLTVGDVVTRSEDGTLSVQKQTVLVDEDGSKYGISYKVSEDGERLVGVDYDPVSEKTP 465
Query 64 VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 123
VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF
Sbjct 466 VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 525
Query 124 DELLMPEFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVF 183
DELLMPEFIGGISRELSMRTVEQT+DQQ A SQGQYAEALGSKTGIAGVYGSTSNNIEVF
Sbjct 526 DELLMPEFIGGISRELSMRTVEQTVDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVF 585
Query 184 CDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVG 243
CDEESYIIGLLTVTPVP+Y+QLLPKDF YNGLLDHYQPEFDRIGFQPITYKE+CP+N
Sbjct 586 CDEESYIIGLLTVTPVPIYSQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNF-D 644
Query 244 DDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSD 303
+ ++KTFGYQRPWYEYVAKYD+AHGLFR D+K F+M R F GLPQLGQQFLLVD D
Sbjct 645 ISNMNDMNKTFGYQRPWYEYVAKYDNAHGLFRTDLKKFVMHRTFSGLPQLGQQFLLVDPD 704
Query 304 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 341
TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 705 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 304 bits (778), Expect = 1e-97, Method: Compositional matrix adjust.
Identities = 152/338 (45%), Positives = 219/338 (65%), Gaps = 14/338 (4%)
Query 4 FVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 63
VGLT + V+ D G T +VDEDG+ Y V ++ +GE L GV+Y P+
Sbjct 457 LVGLTTYESVSLNDAGHTVTTVNTAIVDEDGNAYKVDFE--SNGEALKGVNYTPLKAGE- 513
Query 64 VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 123
AIN + +L + SG +I R VNAYQ++LELN +GFSYK+I++GR+D+++R+
Sbjct 514 --AIN----MQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRY 567
Query 124 DELLMPEFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVF 183
D L MPE++GGI+R++ + + QT++ G Y +LGS+ G+A +G++ +I VF
Sbjct 568 DALNMPEYLGGITRDIVVNPITQTVETT---DTGSYVGSLGSQAGLATCFGNSDGSISVF 624
Query 184 CDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVG 243
CDEES ++G++ V P+PVY +LPK Y LD + PEFD IG+QPI KE+ +
Sbjct 625 CDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAF- 683
Query 244 DDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSD 303
+ + L+ FGYQRPWYEYV K D AHGLF ++NFIM R F+ P+LG+ F ++
Sbjct 684 -ESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPG 742
Query 304 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 341
+VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 743 SVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 780
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 118 bits (296), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 100/337 (30%), Positives = 154/337 (46%), Gaps = 45/337 (13%)
Query 5 VGLTVGDVVTRAD---DGTYSIQKQTVLVDEDGSKYGVSYKVSED---GERLVGVDYDPV 58
V + +GD+ +D DG S+ K E+ + VS V+ D GE VG D DP
Sbjct 199 VDIPLGDITLNSDWYLDG--SVPKL-----ENSTGGYVSGNVTSDNATGEINVGSDNDPF 251
Query 59 SEKTPVTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWD 118
A + L +T TI LR Q++LEL R G Y + ++ +D
Sbjct 252 -------AYDPDGSLEVTST------TITDLRRAFKLQEWLELLARGGSRYIEQIKTFFD 298
Query 119 IDIRFDELLMPEFIGGISRELSMRTVEQTIDQQGADSQGQYA-EALGSKTGIAGVYGSTS 177
+ L PE+I G + + + + T + QG + + TG G Y
Sbjct 299 VKSSDQRLQRPEYITGTKQPVIISEILNTTGETAGLPQGNMSGHGVAVGTGNVGKY---- 354
Query 178 NNIEVFCDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEIC 237
+C+E +IIG+L+VTP Y Q +PK ++ D Y P+F IG QPI EI
Sbjct 355 -----YCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEIY 409
Query 238 PMNIVGDDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQF 297
+ GD+ TFGY + EY G FR + + +R+F LP L Q F
Sbjct 410 AYDPTGDE-------TFGYTPRYAEYKYMPSRVAGEFRSSLDEWHAARIFSSLPTLSQDF 462
Query 298 LLVDSDTVNQVFSVTEYTDKIFGYV--KFNATARLPI 332
+ VD+D +++F+VT+ D ++ +V K A ++P+
Sbjct 463 IEVDADDFDRIFAVTDGDDNLYMHVLNKVKARRQMPV 499
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 116 bits (291), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 85/267 (32%), Positives = 126/267 (47%), Gaps = 19/267 (7%)
Query 68 NSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELL 127
NS A LA S +I LR Q++LE N R G Y +I+ + + L
Sbjct 265 NSDHLFADLAGATAS--SINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQ 322
Query 128 MPEFIGGISRELSMRTVEQT-IDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDE 186
PEF+GG + +++ V QT + QG A GV +SN + C+E
Sbjct 323 RPEFLGGSATPITISEVLQTSANNTEPTPQGNMAGH--------GVSVGSSNYVSYKCEE 374
Query 187 ESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDD 246
YIIG+++V P Y Q +PK F D+Y P F IG QPI +E+ N D
Sbjct 375 HGYIIGIMSVMPKTAYQQGIPKHFKKLDKFDYYWPSFANIGEQPILNEELYHQNNATD-- 432
Query 247 TEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVN 306
++TFGY + EY + HG FR + + M R+F P L Q F+ ++D V
Sbjct 433 ----TETFGYTPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFGSKPTLNQDFIECNADDVE 488
Query 307 QVFSVTEYTDKIFGYV--KFNATARLP 331
+VF+VT + ++ Y+ + AT +P
Sbjct 489 RVFAVTAGQEHLYVYLHNEVKATRLMP 515
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 102 bits (253), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 78/262 (30%), Positives = 120/262 (46%), Gaps = 27/262 (10%)
Query 85 TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV 144
TI LR Q+FLE N R G Y + ++ + + L PE+I G + ++ +
Sbjct 263 TINDLRRAMRLQEFLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEI 322
Query 145 EQTIDQQGADSQGQYAEALGSKTGIA-----GVYGSTSNNIEVFCDEESYIIGLLTVTPV 199
T+ +G + E G+ TG A G YG+ S +E YIIG++++ P
Sbjct 323 TSTV--KGIE------EPQGNPTGKATSLQSGHYGNYS------VEEHGYIIGIMSIMPK 368
Query 200 PVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRP 259
P Y Q +PK ++ +LD Y P F IG Q + +EI T + FGY
Sbjct 369 PAYQQGIPKSYLKTDVLDFYWPSFANIGEQEVLQEEIY-------GYTPTPNAVFGYIPR 421
Query 260 WYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTE-YTDKI 318
+ EY + GLFR + + + R+F +P L Q F+ V ++F+ E Y D I
Sbjct 422 YAEYKYLQNRVCGLFRTTLDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNI 481
Query 319 FGYVKFNATARLPISRVAIPRL 340
+ +V TAR P+ P L
Sbjct 482 YIHVLNKMTARRPMPVFGTPML 503
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 96.7 bits (239), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 70/256 (27%), Positives = 113/256 (44%), Gaps = 20/256 (8%)
Query 85 TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV 144
TI LR Q++LE N R G Y + + + + L PE+I G+ + + V
Sbjct 268 TINDLRRAFKLQEWLEKNARGGTRYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEV 327
Query 145 EQTIDQQGADSQGQYA-EALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVYT 203
T Q G QG A + +G +G Y +C+E YIIG+++V P Y
Sbjct 328 LNTTGQDGGLPQGNMAGHGISVTSGKSGSY---------YCEEHGYIIGIMSVMPKTAYQ 378
Query 204 QLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYEY 263
Q +P+ F+ LD++ P F IG Q + +E+ +D TFGY + EY
Sbjct 379 QGIPRTFLKTDSLDYFWPTFANIGEQEVAKQELYAYTANAND-------TFGYVPRYAEY 431
Query 264 VAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTE-YTDKIFGYV 322
G FR + + + R+F P L F ++ D ++F+V + TD ++ +V
Sbjct 432 KYMPSRVAGEFRTSLNYWHLGRIFATEPSLNSDF--IECDPTKRIFAVEDPETDVLYCHV 489
Query 323 KFNATARLPISRVAIP 338
A P+ + P
Sbjct 490 LNKIKAVRPMPKYGTP 505
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 94.4 bits (233), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 71/262 (27%), Positives = 116/262 (44%), Gaps = 17/262 (6%)
Query 80 QGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISREL 139
Q I T+R Q++LE N R G Y + + + + L PE++GG +
Sbjct 273 QADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGGTKGNM 332
Query 140 SMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPV 199
+ V T + A+ A+G+ G G+ N I+ C+E +IIG++ V PV
Sbjct 333 VISEVLATAETTDANV------AVGTMAG-HGITAHGGNTIKYRCEEHGWIIGIINVQPV 385
Query 200 PVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRP 259
Y Q LP++F LD+ P F IG Q + KE+ + D+ FGY
Sbjct 386 TAYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKELYANSPAPDE-------IFGYIPR 438
Query 260 WYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYT-DKI 318
+ E K G R + + + R+F P L ++F+ + DT ++F+VT+ D I
Sbjct 439 YAEMKFKNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECNPDT--RIFAVTDPAEDHI 496
Query 319 FGYVKFNATARLPISRVAIPRL 340
++ T + R IP +
Sbjct 497 VAHIYNEVTVNRKLPRYGIPTI 518
> Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1
Length=545
Score = 92.0 bits (227), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 69/260 (27%), Positives = 115/260 (44%), Gaps = 25/260 (10%)
Query 85 TIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTV 144
TI LR A Q++ E R G Y++ +Q WD+ I + +PE++GG ++M +
Sbjct 296 TINDLRKAVAVQQYYEALARGGSRYREQVQALWDVVISDKTMQIPEYLGGGRYHVNMNQI 355
Query 145 EQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFC---DEESYIIGLLTVTPVPV 201
QT QQ D + G G T N F +E ++IG+ V
Sbjct 356 VQTSGQQTNDD---------TPIGETGAMSVTPINESSFTKSFEEHGFVIGVCCVRHNHS 406
Query 202 YTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWY 261
Y Q L + + LD+Y P+F +G QPI KEI D++ TFGYQ W
Sbjct 407 YQQGLERFWSRTDRLDYYVPQFANLGEQPIKKKEIMLTGTASDEE------TFGYQEAWA 460
Query 262 EYVAKYDSAHGLFRKDMKN----FIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTDK 317
+Y K + GL R + + + + + +P L Q+++ + + + + + +
Sbjct 461 DYRMKPNRVSGLMRSNAEGTLDFWHYADNYSTVPTLSQEWIAEGKEEIARTL-IVQNEPQ 519
Query 318 IFGYVKF--NATARLPISRV 335
FG ++ T R+P+ V
Sbjct 520 FFGAIRIANKTTRRMPLYSV 539
> Gokush_gi|9629155|ref|NP_044312.1|_VP1_[Chlamydia_phage_1]
Length=596
Score = 82.4 bits (202), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 73/280 (26%), Positives = 123/280 (44%), Gaps = 27/280 (10%)
Query 70 YAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMP 129
Y +LAA S TI +LR Q++ E + R G Y + +QG + + + P
Sbjct 332 YVDLAA-----SSSVTINSLRNAITLQQWFEKSARYGSRYVESVQGHFGVHLGDYRAQRP 386
Query 130 EFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCD---E 186
++GG +S+ V Q QG + Y +++ +F E
Sbjct 387 IYLGGSKSYVSVNPVVQNSSTDSVSPQGNLS-----------AYALSTDTKHLFTKSFVE 435
Query 187 ESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEI-CPMNIVGDD 245
++IGLL+ T Y Q L + + D+Y P F +G QP+ KEI C + V D
Sbjct 436 HGFVIGLLSATADLTYQQGLERQWSRFSRYDYYWPTFAHLGEQPVYNKEIYCQSDTVMDP 495
Query 246 DTEQLSKT-FGYQRPWYEYVAKYDSAHGLFRKD----MKNFIMSRVFKGLPQLGQQFLLV 300
++ FGYQ + EY K GLFR + + ++ +S+ F LP L + F+
Sbjct 496 SGSAVNDVPFGYQERYAEYRYKPSKVTGLFRSNATGTLDSWHLSQNFANLPTLNETFIQS 555
Query 301 DSDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 340
++ +++ +V + D I + FN P+ ++P L
Sbjct 556 NT-PIDRALAVPDQPDFICDFY-FNYRCIRPMPVYSVPGL 593
Lambda K H a alpha
0.319 0.138 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 29312352