bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_2
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
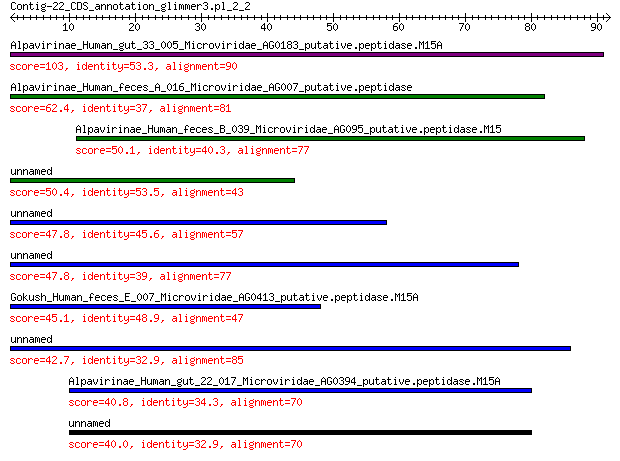
Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.pepti... 103 4e-31
Alpavirinae_Human_feces_A_016_Microviridae_AG007_putative.pepti... 62.4 2e-15
Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.pepti... 50.1 4e-11
unnamed protein product 50.4 4e-11
unnamed protein product 47.8 3e-10
unnamed protein product 47.8 4e-10
Gokush_Human_feces_E_007_Microviridae_AG0413_putative.peptidase... 45.1 3e-09
unnamed protein product 42.7 2e-08
Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.pepti... 40.8 9e-08
unnamed protein product 40.0 2e-07
> Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.peptidase.M15A
Length=131
Score = 103 bits (258), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 48/90 (53%), Positives = 65/90 (72%), Gaps = 3/90 (3%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
+R+ +GKPI + SG+R+ F+N VGGVS SQH G AAD+ D LF LI + +L
Sbjct 44 VREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRC---DDPKALFDLIAESDL 100
Query 61 PFDQVIYYRKAGFIHVSWSPTYRKQIIVRD 90
PFDQ+IYY+K F+HVS+SPTYR ++IV+D
Sbjct 101 PFDQLIYYQKKKFVHVSYSPTYRHEVIVKD 130
> Alpavirinae_Human_feces_A_016_Microviridae_AG007_putative.peptidase
Length=109
Score = 62.4 bits (150), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 43/81 (53%), Gaps = 0/81 (0%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
+R GKPI +NSGYR +NE VGGV S H G AAD ++ I+ +F +++ +
Sbjct 26 VRAHIGKPILVNSGYRCKRLNEMVGGVQKSMHTKGLAADFRTGKKEDINTMFEFLKENQK 85
Query 61 PFDQVIYYRKAGFIHVSWSPT 81
F + FIH+ S T
Sbjct 86 EFKIIELINYKTFIHMGVSET 106
> Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.peptidase.M15
Length=102
Score = 50.1 bits (118), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 31/80 (39%), Positives = 42/80 (53%), Gaps = 5/80 (6%)
Query 11 INSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKME---LPFDQVIY 67
+ S R+ N+A GGV NSQH G A DI + ++L +I PFDQ+I
Sbjct 21 VTSARRTPEQNKAAGGVPNSQHLVGEAVDIKPYGSTTFNKLLEMIHFFSDNVSPFDQLII 80
Query 68 YRKAGFIHVSWSPTYRKQII 87
Y FIHVS+ R+Q+I
Sbjct 81 Y--PTFIHVSFCSRNRRQVI 98
> unnamed protein product
Length=130
Score = 50.4 bits (119), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVS 43
+R F KP+ + SGYR+ N +GGV NSQH G AADI VS
Sbjct 47 IRAHFNKPVIVTSGYRTPEYNTKIGGVKNSQHTKGTAADIKVS 89
> unnamed protein product
Length=128
Score = 47.8 bits (112), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDS--ISELFALIQK 57
+R+ F +P+ INSGYR+ N+ VGG NS H G AADI V S ++E + I K
Sbjct 45 IREHFNEPVIINSGYRTPAWNKKVGGAENSYHCKGMAADIRVKGHTSKEVAEYASKIMK 103
> unnamed protein product
Length=147
Score = 47.8 bits (112), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 30/79 (38%), Positives = 41/79 (52%), Gaps = 4/79 (5%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVS--SRDSISELFALIQKM 58
LR G + + SG+R+ VN +VGGV NS H +G AADI V + + I +
Sbjct 48 LRSQLGCAVIVTSGFRNVSVNNSVGGVLNSDHLYGLAADIRVKGYTPTKLCNFIRSIPLL 107
Query 59 ELPFDQVIYYRKAGFIHVS 77
+ QVI Y F+HVS
Sbjct 108 NVQVGQVIIY--PTFLHVS 124
> Gokush_Human_feces_E_007_Microviridae_AG0413_putative.peptidase.M15A
Length=127
Score = 45.1 bits (105), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 23/47 (49%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDS 47
+R+ F P+ INSGYR+ N VGG NS H G AADI V S
Sbjct 45 IRNHFDAPVIINSGYRTPSWNAKVGGAPNSYHCKGMAADIVVKGHSS 91
> unnamed protein product
Length=130
Score = 42.7 bits (99), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/85 (33%), Positives = 42/85 (49%), Gaps = 3/85 (4%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
+R++ P+ INSGYR+ N VGG S H G AADI+V + E+ + ++
Sbjct 46 IRESVNAPVIINSGYRTPEWNAKVGGAKCSYHVKGMAADIAVKGH-TTKEVAEIASRILG 104
Query 61 PFDQVIYYRKAGFIHVSWSPTYRKQ 85
VI Y F+HV Y ++
Sbjct 105 NHGGVIRY--TNFVHVDVREGYYRK 127
> Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.peptidase.M15A
Length=102
Score = 40.8 bits (94), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 38/70 (54%), Gaps = 3/70 (4%)
Query 10 CINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMELPFDQVIYYR 69
+ S +R+ NEA G +SQH G A D+ S+ L ++I+ FDQ+I YR
Sbjct 20 TVTSAFRTKEQNEACNGSDHSQHLTGDAIDLKPLDF-SVDLLVSMIKGSSFKFDQLIKYR 78
Query 70 KAGFIHVSWS 79
F+H+S++
Sbjct 79 --TFVHISFA 86
> unnamed protein product
Length=102
Score = 40.0 bits (92), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/70 (33%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 10 CINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMELPFDQVIYYR 69
+ S +R+ N+A G SQH G A D+ S+ +I+ FDQ+I YR
Sbjct 20 TVTSAFRTEEQNDACNGSKTSQHLTGDAIDLKPVD-SSVDGFLLMIKGSPFKFDQIIKYR 78
Query 70 KAGFIHVSWS 79
F+HVS++
Sbjct 79 T--FVHVSFA 86
Lambda K H a alpha
0.323 0.138 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4169376