bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
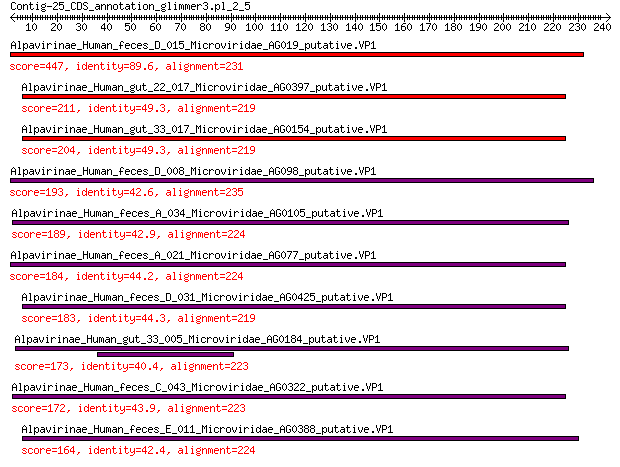
Query= Contig-25_CDS_annotation_glimmer3.pl_2_5
Length=242
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 447 6e-157
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 211 2e-65
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 204 9e-63
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 193 1e-58
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 189 3e-57
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 184 1e-55
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 183 3e-55
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 173 1e-51
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 172 2e-51
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 164 3e-48
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 447 bits (1150), Expect = 6e-157, Method: Compositional matrix adjust.
Identities = 207/231 (90%), Positives = 220/231 (95%), Gaps = 0/231 (0%)
Query 1 MSLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPV 60
M LFNLSSVKN PRRSGFDLSSKVAF+AKVGELLP+KWTLTMPGDKFSLKEQHFTRTQPV
Sbjct 1 MGLFNLSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQPV 60
Query 61 NTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQ 120
NTSAYTRVREYYDWFWCPLHLLWRNAPEVI+QIQQNVQHASSFDGSVLLGSNMPC SADQ
Sbjct 61 NTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSFDGSVLLGSNMPCFSADQ 120
Query 121 ISQSLDQLKSKQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYNQ 180
ISQSLD +KSK NYFGF+RADLAYKL+QYLRYGNVRTGVG++G+RNYGTS+DVKD +YNQ
Sbjct 121 ISQSLDMMKSKLNYFGFNRADLAYKLIQYLRYGNVRTGVGTSGSRNYGTSVDVKDSSYNQ 180
Query 181 NRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKGCCYYL 231
NRA+NHALS+FPILAYKKFCQDYFRLTQWQ SAPYLWNIDYYDGKG L
Sbjct 181 NRAFNHALSVFPILAYKKFCQDYFRLTQWQVSAPYLWNIDYYDGKGATTIL 231
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 211 bits (536), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 108/230 (47%), Positives = 153/230 (67%), Gaps = 12/230 (5%)
Query 6 LSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
+S+++NHP RSGFD+ K AFTAKVGELLPV W ++MPGDK+ ++FTRTQPV TSAY
Sbjct 1 MSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYKFNVEYFTRTQPVETSAY 60
Query 66 TRVREYYDWFWCPLHLLWRNAPEVISQIQQ-NVQHASSFDGSVLLGSNMPCLSADQISQS 124
TR+REY+D++ PL LLW++AP V++Q+Q N A SF ++ LGS P L+ + +
Sbjct 61 TRLREYFDFYAVPLRLLWKSAPSVLTQMQDINQLQALSFTQNLSLGSYFPSLTLSRFTAV 120
Query 125 LDQLKSKQNY----------FGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVK 174
L++L N FGF RADLA+KL YL YGNV + S+ R + TS+
Sbjct 121 LNRLNGGSNVPGNSSTFLNEFGFSRADLAFKLFSYLGYGNVWSSEFSSSNRWWSTSLK-G 179
Query 175 DGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDG 224
G+Y Q + +++FP+LAY+K QD+FR +QW++S P +N+DY+ G
Sbjct 180 GGSYTQQYVQDSYVNLFPLLAYQKIYQDFFRWSQWENSNPSSYNVDYFTG 229
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 204 bits (518), Expect = 9e-63, Method: Compositional matrix adjust.
Identities = 108/232 (47%), Positives = 150/232 (65%), Gaps = 13/232 (6%)
Query 6 LSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
LS+++NHP RSGFD+ K AFTAKVGELLPV W ++MPGDK+ ++FTRTQPV TSAY
Sbjct 7 LSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNVEYFTRTQPVATSAY 66
Query 66 TRVREYYDWFWCPLHLLWRNAPEVISQIQQ-NVQHASSFDGSVLLGSNMPCLSADQISQS 124
TR+REY+D++ PL LLW++AP V++Q+Q N A S ++ LG+ +P L+ + +
Sbjct 67 TRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLTIGTLGWA 126
Query 125 LDQLKSK----------QNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSI-DV 173
+ L +N FGF RADL++KLL YL YGN+ S G R + TS+ +
Sbjct 127 IRYLNGNTWEPETASYLRNAFGFSRADLSFKLLSYLGYGNLIETPPSLGNRWWSTSLKNT 186
Query 174 KDGT-YNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDG 224
DG Y Q N ++IFP+L Y+K QD+FR QW+ S P +N+DY+ G
Sbjct 187 DDGANYTQQYIQNTIVNIFPLLTYQKIYQDFFRWPQWEKSNPSSYNVDYFSG 238
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 193 bits (490), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 100/249 (40%), Positives = 151/249 (61%), Gaps = 16/249 (6%)
Query 1 MSLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPV 60
MSLFN+S+VKNHPRRSGFDLS++V FT+K GELLPV W + PGD F +K Q FTRTQP+
Sbjct 1 MSLFNMSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPL 60
Query 61 NTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQ 120
NT+AYTR+REY D+++ PL L+ +N P ++Q+Q N A+ + ++ +++P +
Sbjct 61 NTAAYTRIREYLDFYFVPLRLINKNLPTALTQMQDNPVQATGLSSNKVVTTDIPWVPVHS 120
Query 121 ISQSLDQLKS------------KQNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYG 168
S S L +N+ GFD + KLL YLRYGN + V + ++ G
Sbjct 121 SSGSYSSLTGFADVRGSVSSSDIENFLGFDSITQSAKLLMYLRYGNFLSSVVPDEQKSIG 180
Query 169 --TSIDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGKG 226
+S+D+++ + + Y ++ I P+ Y+K D+FR TQW+ + PY +N D+Y G
Sbjct 181 LSSSLDLRN-SETVSTGYT-SVHILPLATYQKIYADFFRFTQWEKNQPYTYNFDWYSGGN 238
Query 227 CCYYLAYKS 235
L ++
Sbjct 239 VLASLTTQA 247
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 189 bits (480), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 96/227 (42%), Positives = 140/227 (62%), Gaps = 25/227 (11%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
SLF+ +KN PRRSGFDLS+K AFTAKVGELLPV W +PGDKF++ ++ FTRTQPV+
Sbjct 3 SLFSYGDIKNSPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQPVD 62
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQI 121
TSA+TR+REYY+WF+ PLHLL+RN+ E I ++ +A+S S+ N+P + I
Sbjct 63 TSAFTRIREYYEWFFVPLHLLYRNSNEAIMSMENQPNYAASGSSSISFNRNLPWVDLSTI 122
Query 122 SQSLDQLKSK---QNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTY 178
+ ++ ++S +N+FG R++ KL+ YL YG T
Sbjct 123 NVAIGNVQSSTSPKNFFGVSRSEGFKKLVSYLGYGE----------------------TS 160
Query 179 NQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGK 225
+ N S FP+ AY+K QDY+R +QW+ S P+ +N D+++G+
Sbjct 161 PEKYVDNLRCSAFPLYAYQKIYQDYYRHSQWEKSKPWTYNCDFWNGE 207
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 184 bits (468), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 99/235 (42%), Positives = 141/235 (60%), Gaps = 15/235 (6%)
Query 1 MSLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPV 60
M LF+ +KNHPRRSGFDL+ ++ FT+K GELLPV W T+PGDK+++ FTRTQPV
Sbjct 1 MGLFSYDDLKNHPRRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPV 60
Query 61 NTSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQ 120
+T+AYTRV+EY DWF+ PL+L+ + I+Q+ N A S + + ++MP +
Sbjct 61 DTAAYTRVKEYVDWFFVPLNLIQKGIESAITQMVDNPVSAMSAIENRAITTDMPYTTLLS 120
Query 121 ISQSLDQLKSKQ---------NYFGFDRADLAYKLLQYLRYGNVRTGVGSN-GARNYG-T 169
+S++L L K N FGF RADL+ KLLQ L+YGN S +G +
Sbjct 121 LSRALYMLNGKSYVNSHAGKLNMFGFSRADLSAKLLQMLKYGNFINPEHSGLDTPMFGYS 180
Query 170 SIDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDG 224
++ + +Y +N A+++ PI Y+K DYFR QW+ PY +N DYY G
Sbjct 181 TVKLAQFSY----LWNQAVNVLPIFCYQKIYSDYFRFQQWEKPVPYTYNADYYAG 231
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 183 bits (465), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 97/234 (41%), Positives = 144/234 (62%), Gaps = 17/234 (7%)
Query 6 LSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
+S+VKNHPRRSGFDLS++V FT+K GELLPV W + PGD F +K Q FTRTQP+NT+AY
Sbjct 1 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 60
Query 66 TRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQISQ-- 123
TR+REY D+++ PL L+ +N P + Q+Q N A+ + ++ +++P + +
Sbjct 61 TRIREYLDFYFVPLRLINKNLPTALMQMQDNPVQATGLSSNKVVTTDIPWVPTNLSGTYG 120
Query 124 SLDQLKSKQNYF-----------GFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGT--S 170
SL L +N F GFD + KLL YLRYGN + V S+ ++ G S
Sbjct 121 SLTALADVKNSFPSSSTGIEDLLGFDAITQSAKLLMYLRYGNFLSSVVSDKNKSLGLSGS 180
Query 171 IDVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDG 224
+D+++ + + Y ++ I P+ AY+K D+FR TQW+ + PY +N D+Y G
Sbjct 181 LDLRN-SETASTGYT-SMHILPLAAYQKAYADFFRFTQWEKNQPYTYNFDWYSG 232
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 173 bits (438), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 90/226 (40%), Positives = 133/226 (59%), Gaps = 25/226 (11%)
Query 3 LFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNT 62
LF+ +KN PRRSGFDL +K AFTAKVGELLPV W MPGDKF + ++ FTRTQPV+T
Sbjct 4 LFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCMPGDKFHISQEWFTRTQPVDT 63
Query 63 SAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQIS 122
SA+TR+REYY+WF+ PLHL++RN+ E I ++ +A+S S++ +P + ++
Sbjct 64 SAFTRIREYYEWFFVPLHLMYRNSNEAIMSLENQPNYAASGTQSIVFNRKLPWVDLQTLN 123
Query 123 QSLDQLKSK---QNYFGFDRADLAYKLLQYLRYGNVRTGVGSNGARNYGTSIDVKDGTYN 179
++ +++ N FGF R+ YKL L GVG T
Sbjct 124 DAITNVQASTYHNNMFGFARSSGFYKLFNSL-------GVGE---------------TDP 161
Query 180 QNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGK 225
N +S FP AY+K D++R +QW+ + P+ +N D+++G+
Sbjct 162 SKTLANLRISAFPFYAYQKIYSDHYRNSQWEVNKPWTYNCDFWNGE 207
Score = 21.6 bits (44), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 11/61 (18%), Positives = 30/61 (49%), Gaps = 6/61 (10%)
Query 36 VKWTLTMPGDKFSLKE------QHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEV 89
+++ T+ GD + + E + +NT+A ++E + + P ++ W+ + +V
Sbjct 472 LQFMTTVDGDSWPVPEMDSIGFEELPSYSLLNTNAVQPIKEPRPFGYVPRYISWKTSVDV 531
Query 90 I 90
+
Sbjct 532 V 532
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 172 bits (437), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 98/232 (42%), Positives = 141/232 (61%), Gaps = 14/232 (6%)
Query 2 SLFNLSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVN 61
+LF+ SVKN P RSGFD S +FTAK GELLPV W + +PG K +L FTRT PVN
Sbjct 3 NLFSYGSVKNKPARSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMPVN 62
Query 62 TSAYTRVREYYDWFWCPLHLLWRNAPEVISQIQQNVQHASSFDGSVLLGSNMPCLSADQI 121
T+AYTRV+EYYDW++ PL L+ ++ + + Q+Q A+S + + ++P +A +
Sbjct 63 TAAYTRVKEYYDWYFVPLRLINKSIGQALVQMQDQPVQATSIVANKSVTLDLPWTNAATM 122
Query 122 SQSLD----QLKSKQNYFGFDRADLAYKLLQYLRYGNV-----RTGVGSNGARNYGTSID 172
L+ L +K N GF +A + KLL+YLRYGN + VG N +N+G S
Sbjct 123 FTLLNYANVILTNKYNLDGFSKAATSAKLLRYLRYGNCYYTSDPSKVGKN--KNFGLS-- 178
Query 173 VKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDG 224
KD +N A N + ++ P+ AY+K D+FR QW+++ PY +N DYY+G
Sbjct 179 SKDD-FNLLAAKNVSFNVLPLAAYQKIYCDWFRFEQWENACPYTYNFDYYNG 229
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 164 bits (414), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 95/244 (39%), Positives = 144/244 (59%), Gaps = 21/244 (9%)
Query 6 LSSVKNHPRRSGFDLSSKVAFTAKVGELLPVKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
L ++NHP ++GFD+ SK FTAKVGEL+PV W +P + + +FTRT+PV T+AY
Sbjct 7 LKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRPVQTAAY 66
Query 66 TRVREYYDWFWCPLHLLWRNAPEVISQIQQN--VQHASSFDGSVLLGSNMPCLSADQISQ 123
TRVREY+D++ P LLW++ + Q+ Q VQ + D + +G+++P + +S
Sbjct 67 TRVREYFDFYAVPCDLLWKSFDSAVIQMGQTAPVQSKTLLD-PLTVGNDIPWCTLLDLSN 125
Query 124 SL------DQLKSK-------QNYFGFDRADLAYKLLQYLRYGNV----RTGVGSNGARN 166
++ L S N FG++R D+ KLL YL YGN + VGS R
Sbjct 126 AVYFSSGSSPLGSTVSVPSGFGNIFGYNRGDVDSKLLFYLNYGNFVNPSLSNVGSPSNRW 185
Query 167 YGTSI-DVKDGTYNQNRAYNHALSIFPILAYKKFCQDYFRLTQWQDSAPYLWNIDYYDGK 225
+ TS K Y+Q N+A++IFP+LAY+K QD+FR +QW+++ P +N+DYY+G
Sbjct 186 WNTSFSSSKVPGYSQKYLNNNAVNIFPLLAYQKIYQDFFRWSQWENADPTSYNVDYYNGS 245
Query 226 GCCY 229
G +
Sbjct 246 GNLF 249
Lambda K H a alpha
0.322 0.136 0.438 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 19270416