bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_3
Length=222
Score E
Sequences producing significant alignments: (Bits) Value
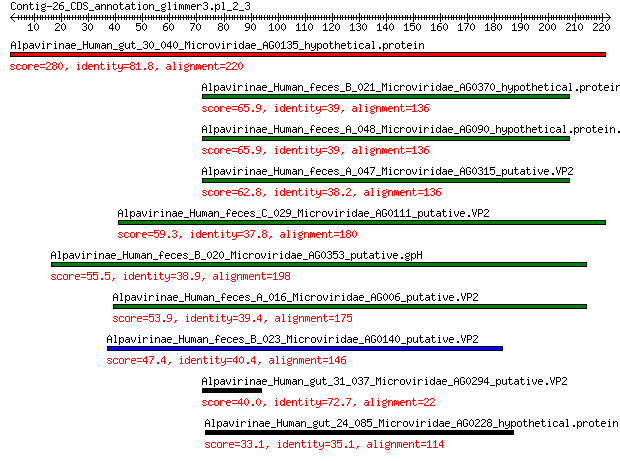
Alpavirinae_Human_gut_30_040_Microviridae_AG0135_hypothetical.p... 280 1e-94
Alpavirinae_Human_feces_B_021_Microviridae_AG0370_hypothetical.... 65.9 1e-14
Alpavirinae_Human_feces_A_048_Microviridae_AG090_hypothetical.p... 65.9 1e-14
Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2 62.8 1e-13
Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2 59.3 3e-12
Alpavirinae_Human_feces_B_020_Microviridae_AG0353_putative.gpH 55.5 5e-11
Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2 53.9 2e-10
Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2 47.4 2e-08
Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2 40.0 8e-06
Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.p... 33.1 0.001
> Alpavirinae_Human_gut_30_040_Microviridae_AG0135_hypothetical.protein
Length=377
Score = 280 bits (717), Expect = 1e-94, Method: Compositional matrix adjust.
Identities = 180/220 (82%), Positives = 201/220 (91%), Gaps = 1/220 (0%)
Query 1 MGNFKESFISGLGSTAASSGIGFIGNILSQALGLSWSPQKAMEEQWKYNKKIMalqnqyq 60
MG F +SFI+GLGSTAAS+GIGFIGN LSQA GLSWSP++AM+EQ YNK+IMALQNQYQ
Sbjct 1 MG-FGKSFINGLGSTAASTGIGFIGNALSQAFGLSWSPERAMKEQEAYNKRIMALQNQYQ 59
Query 61 qqaaaqsqqyaKDYWDYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQP 120
QQAAAQSQQYAKDYWDYTN ENQV+HLKN+GLN+GLMYGQSGAGGMGA+GGA Q SP+Q
Sbjct 60 QQAAAQSQQYAKDYWDYTNAENQVRHLKNSGLNIGLMYGQSGAGGMGASGGARQDSPDQA 119
Query 121 QGNPVGMALQVQQIEQQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQ 180
QGNP+GMALQVQQ+EQQRRMNDAQIALAEAQA+KA EA KI+GVDTQEALKRIE S+
Sbjct 120 QGNPIGMALQVQQLEQQRRMNDAQIALAEAQANKAGAEANKIAGVDTQEALKRIEEAGSR 179
Query 181 IELNLKEGQYKEALTDLTKAEKEATEALTSLREMQEGLTR 220
IELNLKE YK+ALTDLTKAEKEAT+AL +LREMQEGLT+
Sbjct 180 IELNLKESNYKDALTDLTKAEKEATDALKALREMQEGLTK 219
> Alpavirinae_Human_feces_B_021_Microviridae_AG0370_hypothetical.protein.BACPLE.00802
Length=333
Score = 65.9 bits (159), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/136 (39%), Positives = 75/136 (55%), Gaps = 2/136 (1%)
Query 72 KDYWDYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQPQGNPVGMALQV 131
K+YWDYTN EN VKHLK AGLN L Y + G GG G A P P +Q
Sbjct 29 KNYWDYTNYENSVKHLKEAGLNPALFYAKGGQGGATGGGQAQGVGL--PSTTPTMARIQA 86
Query 132 QQIEQQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYK 191
Q + Q + +Q+ L +A A K EA+KI+G DT+ A + E + SQ E N + + +
Sbjct 87 QGMGAQLQNVLSQVELNKATAKKTEAEAEKIAGADTKVAEREAEMLESQSEFNKRVTKLQ 146
Query 192 EALTDLTKAEKEATEA 207
+++ L KA+++ T A
Sbjct 147 DSIEKLNKAQEQKTAA 162
> Alpavirinae_Human_feces_A_048_Microviridae_AG090_hypothetical.protein.BACPLE.00802
Length=333
Score = 65.9 bits (159), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/136 (39%), Positives = 75/136 (55%), Gaps = 2/136 (1%)
Query 72 KDYWDYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQPQGNPVGMALQV 131
K+YWDYTN EN VKHLK AGLN L Y + G GG G A P P +Q
Sbjct 29 KNYWDYTNYENSVKHLKEAGLNPALFYAKGGQGGATGGGQAQGVGL--PPTTPTMARIQA 86
Query 132 QQIEQQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYK 191
Q + Q + +Q+ L +A A K EA+KI+G DT+ A + E + SQ E N + + +
Sbjct 87 QGMGAQLQNVLSQVELNKATAKKTEAEAEKIAGADTKVAEREAEMLESQSEFNRRITRLQ 146
Query 192 EALTDLTKAEKEATEA 207
+++ LT A+++ T A
Sbjct 147 DSIEKLTNAQEQKTAA 162
> Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2
Length=325
Score = 62.8 bits (151), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 52/136 (38%), Positives = 75/136 (55%), Gaps = 1/136 (1%)
Query 72 KDYWDYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQPQGNPVGMALQV 131
KD WDYTN ENQ +H+KNAGLN + G GG + GA QP V M L+
Sbjct 20 KDLWDYTNYENQKQHIKNAGLN-PALMYGMGGGGGVSANGAQGQGVTQPTDRSVEMKLKQ 78
Query 132 QQIEQQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYK 191
Q + Q +Q+ L ++ A K EA KI+G DT+ A K+ E + SQ E N + + +
Sbjct 79 QGLGLQLASIASQVELNKSLAEKNKVEADKIAGADTKVAEKQAEMLESQSEFNKRITKLQ 138
Query 192 EALTDLTKAEKEATEA 207
+++ LT A+++ T A
Sbjct 139 DSIEKLTNAQEQKTAA 154
> Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2
Length=329
Score = 59.3 bits (142), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 68/194 (35%), Positives = 98/194 (51%), Gaps = 17/194 (9%)
Query 41 AMEEQWKYNKKIMalqnqyqqqaaaqsqqyaKDYWDYTNVENQVKHLKNAGLNVglmygq 100
A EQW K+M +QN+Y +Q A +QQ KD WDYTN ENQ +H++NAGLN +
Sbjct 5 AANEQWGNQLKLMEIQNRYNEQMAKNNQQRNKDLWDYTNYENQKQHIENAGLN-PALMYG 63
Query 101 sgaggmgatggahqaSPEQPQGNPVGMALQVQQIEQQRRMNDAQIALAEAQASKANEEAK 160
G GG + GA QP V M L+ Q + Q +Q+ L ++QA K EA+
Sbjct 64 MGGGGGISANGAQGQGVTQPTDRSVEMGLKQQGLGLQLASIASQVDLNKSQAEKNKAEAE 123
Query 161 KISGVDTQEALKRIEG--------------IASQIELNLKEGQYKEALTDLTKAEKEATE 206
KISGVDT+ I+ I QI + E + K + D TK +A E
Sbjct 124 KISGVDTRAQEATIDNLIAQTSNEKVKKGLILGQIRVADAEEELKRNMADWTK--DKADE 181
Query 207 ALTSLREMQEGLTR 220
+++ +Q+G+ +
Sbjct 182 TRWNIKSLQKGIDK 195
> Alpavirinae_Human_feces_B_020_Microviridae_AG0353_putative.gpH
Length=352
Score = 55.5 bits (132), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 77/198 (39%), Positives = 105/198 (53%), Gaps = 21/198 (11%)
Query 16 AASSGIGFIGNILSQALGLSWSPQKAMEEQWKYNKKIMalqnqyqqqaaaqsqqyaKDYW 75
AA+S IG SQ G+ + A E + K++M Q + A +QQ AK W
Sbjct 8 AATSAIG------SQIKGIE--ARDAEERAYTKQKELMDKQYALNDKMAEANQQRAKYMW 59
Query 76 DYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQPQGNPVGMALQVQQIE 135
DYTN ENQ +HL NA L+ GL YG SGAGG +GG + VG +Q + +
Sbjct 60 DYTNFENQKQHLLNANLSPGLFYGGSGAGGSTTSGGQGSGVGLGTE-TGVGYGIQEKALG 118
Query 136 QQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYKEALT 195
Q +Q+AL ++QA+K N EAKKISGVDTQ SQ +LN + E LT
Sbjct 119 LQLASMASQVALNQSQANKNNAEAKKISGVDTQLT-------ESQTKLN----KAMENLT 167
Query 196 DLTKAEKEATEALTSLRE 213
+ TK ++EA + +L+E
Sbjct 168 N-TKEQREAADYFVALQE 184
> Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2
Length=367
Score = 53.9 bits (128), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 69/175 (39%), Positives = 107/175 (61%), Gaps = 4/175 (2%)
Query 39 QKAMEEQWKYNKKIMalqnqyqqqaaaqsqqyaKDYWDYTNVENQVKHLKNAGLNVglmy 98
+K ME+ W+Y K+ M +Q QY Q AA ++Q+ + W+ TN Q +H+++AGL+VGLMY
Sbjct 56 EKLMEKAWEYEKEGMGMQYQYGQAAANEAQRRNMEMWNQTNFGAQRQHMEDAGLSVGLMY 115
Query 99 gqsgaggmgatggahqaSPEQPQGNPVGMALQVQQIEQQRRMNDAQIALAEAQASKANEE 158
G G G + G A Q P P NPVGMALQ +QIEQQ +Q L +A+A+KA E
Sbjct 116 GGGGQGAVSQGGQATQ--PSGPTSNPVGMALQYKQIEQQNEAIKSQTMLNQAEAAKALAE 173
Query 159 AKKISGVDTQEALKRIEGIASQIELNLKEGQYKEALTDLTKAEKEATEALTSLRE 213
AKK GVDT++ I+ +IE ++E + + A +++ +A+ A + + ++
Sbjct 174 AKKTGGVDTKKTESEIK--WQEIENRIQESREQIASSNIIEAKANAKKTVEEFKQ 226
> Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2
Length=295
Score = 47.4 bits (111), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 59/148 (40%), Positives = 83/148 (56%), Gaps = 12/148 (8%)
Query 37 SPQKAMEEQWKYNKKIMalqnqyqqqaaaqsqqyaKDYWDYTNVENQVKHLKNAGLNVgl 96
S ++AM++QWK K+ MALQ +Y ++ A SQQ A D W+ TN E+QV+H+K AGLN L
Sbjct 3 SQEQAMQDQWKLEKEKMALQAKYNKEQADYSQQLALDMWNATNYESQVEHMKAAGLNPAL 62
Query 97 mygqsgaggmgatggahqaSPEQPQGNPVGMALQVQQ--IEQQRRMNDAQIALAEAQASK 154
+Y + GAGG + G + VGM LQ +Q I Q ++M A+ +K
Sbjct 63 LYSKGGAGGSTSGAGTAAPVSD-GTTQAVGMGLQAKQIAISQAQQM---------AETAK 112
Query 155 ANEEAKKISGVDTQEALKRIEGIASQIE 182
E KISGVDT+ I+ + IE
Sbjct 113 TVAETAKISGVDTESVKTSIKKMLQDIE 140
> Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2
Length=360
Score = 40.0 bits (92), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 16/22 (73%), Positives = 18/22 (82%), Gaps = 0/22 (0%)
Query 72 KDYWDYTNVENQVKHLKNAGLN 93
KD W+YTN ENQV H+K AGLN
Sbjct 60 KDMWNYTNYENQVAHMKAAGLN 81
> Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.protein
Length=335
Score = 33.1 bits (74), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 55/124 (44%), Gaps = 25/124 (20%)
Query 73 DYWDYTNVENQVKHLKNAGLNVglmygqsgaggmgatggahqaSPEQPQGN--------- 123
+ W T E QV+ ++NAGLN GG ++ Q QGN
Sbjct 79 EMWRKTGYEAQVQQIENAGLN-----------EALMYGGGGASATSQSQGNSGVTNTGTQ 127
Query 124 PVGMALQVQQIEQQRRMNDAQIALAEAQASK-ANEEAKKISGVDTQEALKRIEGIASQIE 182
V + LQ + IE Q +A AL AQA+K A E +KK + ++ K I + IE
Sbjct 128 AVALGLQARAIEAQVNNTEADTALKMAQAAKEAGEASKKPIELKIEQTNKEI----ADIE 183
Query 183 LNLK 186
NLK
Sbjct 184 KNLK 187
Lambda K H a alpha
0.308 0.124 0.336 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 17280362