bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_1
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
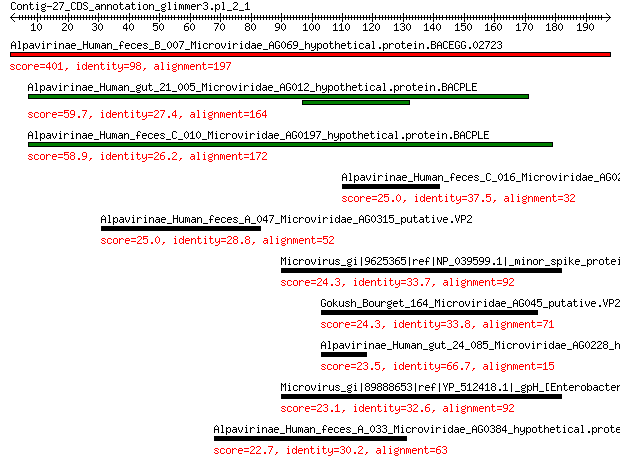
Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.p... 401 2e-142
Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.pr... 59.7 1e-12
Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.... 58.9 2e-12
Alpavirinae_Human_feces_C_016_Microviridae_AG0275_hypothetical.... 25.0 0.39
Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2 25.0 0.48
Microvirus_gi|9625365|ref|NP_039599.1|_minor_spike_protein_[Ent... 24.3 0.67
Gokush_Bourget_164_Microviridae_AG045_putative.VP2 24.3 0.75
Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.p... 23.5 1.6
Microvirus_gi|89888653|ref|YP_512418.1|_gpH_[Enterobacteria_pha... 23.1 2.0
Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.... 22.7 2.4
> Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.protein.BACEGG.02723
Length=383
Score = 401 bits (1030), Expect = 2e-142, Method: Compositional matrix adjust.
Identities = 193/197 (98%), Positives = 196/197 (99%), Gaps = 0/197 (0%)
Query 1 MSGLEVQLTKAQAEYTEGQKSKLISEINDINEHVNLLKAQISETWSRTANLDASTIATRT 60
MSGLEVQLTKAQAEYTE QKS+LISEINDINEHVNLLKAQISETWSRTANLDAST+ATRT
Sbjct 187 MSGLEVQLTKAQAEYTEKQKSRLISEINDINEHVNLLKAQISETWSRTANLDASTVATRT 246
Query 61 AAILNNRRFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIR 120
AAILNNRRFDLECE+FARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIR
Sbjct 247 AAILNNRRFDLECEEFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIR 306
Query 121 LTDAQKTQVEHYTNSIDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSLYHISQVASDW 180
LTDAQKTQVEHYTNSIDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSLYHISQVASDW
Sbjct 307 LTDAQKTQVEHYTNSIDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSLYHISQVASDW 366
Query 181 LPSPGGIAKKLLRHGKK 197
LPSPGGIAKKLLRHGKK
Sbjct 367 LPSPGGIAKKLLRHGKK 383
> Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.protein.BACPLE
Length=383
Score = 59.7 bits (143), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/167 (27%), Positives = 85/167 (51%), Gaps = 13/167 (8%)
Query 7 QLTKAQAEYTEGQKSKLISEINDINEHVNLLKAQISETWSRTANLDASTIATRTAAILNN 66
QL A+A+ + ++ +++ E +N L+AQ SE +D + + L +
Sbjct 197 QLNHAEADLAAKKLQEIDVAMSEARERINTLRAQQSE-------IDEKIVQMKFDRYLRS 249
Query 67 RRFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTAL--KQANIRLTDA 124
+ F+L C + ++E++++++L+ AE + I+ T A+V N++ T + KQ L +
Sbjct 250 KEFELLCVRTYQDIKESNSRISLNAAEVQDIMATQLARVLNLNASTYMMKKQG---LLAS 306
Query 125 QKTQVEHYTNS-IDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSL 170
++T E + + IDI A+F QQ Q +D +R VA SL
Sbjct 307 EQTMTELFKQTGIDISNQQAIFNFQQAQTWDSTERFTNVATTWINSL 353
Score = 23.9 bits (50), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/37 (38%), Positives = 17/37 (46%), Gaps = 2/37 (5%)
Query 97 ILVTMYAKVNNIDTDTALKQANIRLTDAQ--KTQVEH 131
I Y + I DT LK + RL D+Q KT E
Sbjct 128 IPTNTYGQTAQIAADTGLKASQARLADSQSKKTDTEE 164
> Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.protein.BACPLE
Length=382
Score = 58.9 bits (141), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/174 (26%), Positives = 87/174 (50%), Gaps = 10/174 (6%)
Query 7 QLTKAQAEYTEGQKSKLISEINDINEHVNLLKAQISETWSRTANLDASTIATRTAAILNN 66
QL A+AE + ++ +++ E +N +KAQ S+ +D + + + L +
Sbjct 196 QLNHAEAEVAAKKLQEIDVAMSEARERINTMKAQQSQ-------IDENIVQLKFDRYLRS 248
Query 67 RRFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIRLTDAQK 126
F+L C+ + ++E+++++NL+ AE + ++ T A+V N++ T +++ L Q
Sbjct 249 NEFELLCKKTYQDMKESNSRINLNAAEVQDMMATQLARVMNLNASTYMQKKQGMLASEQ- 307
Query 127 TQVEHYTNS-IDIHRDAAVFKLQQDQKYDDAQRIVTVANQATQSL-YHISQVAS 178
T E Y + IDI A F Q + +D +R VA S+ + +SQ A
Sbjct 308 TMTELYKQTGIDISNQHAKFNFDQAKNWDSTERFTNVATTWINSVSFAVSQFAG 361
> Alpavirinae_Human_feces_C_016_Microviridae_AG0275_hypothetical.protein.HMPREF9141.0987
Length=335
Score = 25.0 bits (53), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 12/32 (38%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 110 TDTALKQANIRLTDAQKTQVEHYTNSIDIHRD 141
T+ L++ +RLT AQ T+V D+ RD
Sbjct 245 TEDDLRKGRVRLTGAQTTEVLSMAGLNDVRRD 276
> Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2
Length=325
Score = 25.0 bits (53), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 15/52 (29%), Positives = 25/52 (48%), Gaps = 3/52 (6%)
Query 31 NEHVNLLKAQISETWSRTANLDASTIATRTAAILNNRRFDLECEDFARRVRE 82
NE +N LK QI+ W+ A + +++ + I N + D R+ RE
Sbjct 213 NEQINYLKGQIAIGWANVA-IGEKSVSNESDRIAN--ELMIGIRDLDRKDRE 261
> Microvirus_gi|9625365|ref|NP_039599.1|_minor_spike_protein_[Enterobacteria_phage_alpha3]
Length=330
Score = 24.3 bits (51), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 46/104 (44%), Gaps = 23/104 (22%)
Query 90 SEAEAKSILVTMYAKVNNIDTDTALKQAN-----IRLTDAQKTQVEHYTNSIDIHRDAAV 144
S+A SIL +TD KQA + LT AQ+T +H TNS + + V
Sbjct 216 SQARVASILA---------NTDLTTKQATHEIMRMALTRAQETG-QHLTNSQIMALEKKV 265
Query 145 F----KLQQD---QKYDDAQRIVTVANQATQSLYHISQVASDWL 181
+ K+ QD +Y +Q + A T + + ASDW+
Sbjct 266 YAEIGKIHQDTQNSRYGSSQ-VTAAAKDVTNMITDAASGASDWV 308
> Gokush_Bourget_164_Microviridae_AG045_putative.VP2
Length=261
Score = 24.3 bits (51), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 24/81 (30%), Positives = 38/81 (47%), Gaps = 15/81 (19%)
Query 103 AKVNNIDTDTALKQANIRLTDAQKTQVEHYTNSIDIHRDAAVFKLQQDQKYD-------- 154
A++ N++ DT K+A+ L+ AQK + S D R A L ++Q
Sbjct 115 AEIENVEADTIKKRADTMLSVAQK---DLAGASADEKR--ATINLMENQVNKISAEVKNI 169
Query 155 --DAQRIVTVANQATQSLYHI 173
+ R++ VA Q T+SL I
Sbjct 170 PLEGDRLIAVAKQLTESLPQI 190
> Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.protein
Length=335
Score = 23.5 bits (49), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 10/15 (67%), Positives = 12/15 (80%), Gaps = 0/15 (0%)
Query 103 AKVNNIDTDTALKQA 117
A+VNN + DTALK A
Sbjct 140 AQVNNTEADTALKMA 154
> Microvirus_gi|89888653|ref|YP_512418.1|_gpH_[Enterobacteria_phage_WA13_sensu_lato]
Length=325
Score = 23.1 bits (48), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 30/104 (29%), Positives = 47/104 (45%), Gaps = 23/104 (22%)
Query 90 SEAEAKSILVTMYAKVNNIDTDTALKQA-----NIRLTDAQKTQVEHYTNSIDIHRDAAV 144
S+A +SIL +TD KQA + LT AQ+T +H TNS + + V
Sbjct 211 SQARVESILA---------NTDLTAKQAAHEIMRMALTRAQETG-QHLTNSQIMALEKKV 260
Query 145 F----KLQQD---QKYDDAQRIVTVANQATQSLYHISQVASDWL 181
+ K+ QD +Y +Q + A T + + A+DW+
Sbjct 261 YAEIGKIHQDTQNARYGSSQ-VTAAAKDVTNMITDATSGAADWV 303
> Alpavirinae_Human_feces_A_033_Microviridae_AG0384_hypothetical.protein.BACEGG.02723
Length=367
Score = 22.7 bits (47), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 32/63 (51%), Gaps = 2/63 (3%)
Query 68 RFDLECEDFARRVRETDAKVNLSEAEAKSILVTMYAKVNNIDTDTALKQANIRLTDAQKT 127
R DL + F ++E +++ +EA A + L T AK+ I+ + A+ TDA +
Sbjct 178 REDLTSKRFGNILKEVESRY--AEANAIADLDTKQAKIAEINASVLERLASAAKTDADRI 235
Query 128 QVE 130
VE
Sbjct 236 TVE 238
Lambda K H a alpha
0.313 0.125 0.338 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 14715253