bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_2
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
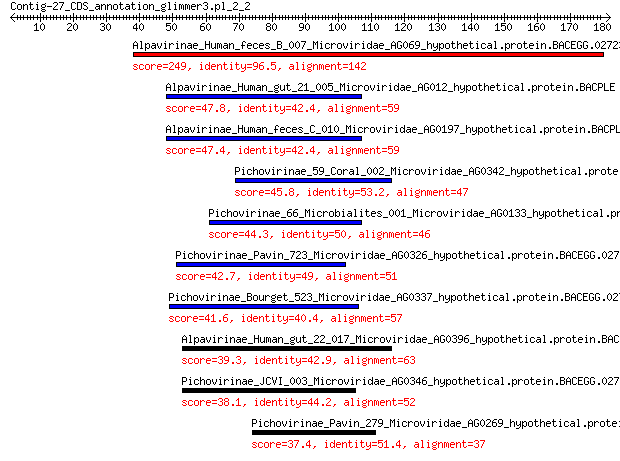
Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.p... 249 3e-83
Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.pr... 47.8 1e-08
Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.... 47.4 1e-08
Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.prot... 45.8 4e-08
Pichovirinae_66_Microbialites_001_Microviridae_AG0133_hypotheti... 44.3 1e-07
Pichovirinae_Pavin_723_Microviridae_AG0326_hypothetical.protein... 42.7 4e-07
Pichovirinae_Bourget_523_Microviridae_AG0337_hypothetical.prote... 41.6 1e-06
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 39.3 9e-06
Pichovirinae_JCVI_003_Microviridae_AG0346_hypothetical.protein.... 38.1 2e-05
Pichovirinae_Pavin_279_Microviridae_AG0269_hypothetical.protein... 37.4 3e-05
> Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.protein.BACEGG.02723
Length=383
Score = 249 bits (636), Expect = 3e-83, Method: Compositional matrix adjust.
Identities = 137/142 (96%), Positives = 139/142 (98%), Gaps = 0/142 (0%)
Query 38 RRQKRAIAAQKEENAQARAWSEKMARWYANNERANLADERAYNNPSAVMSRLKDAGLNPD 97
RRQKRAIAAQKEENAQARAWSEKMARWYANNERANLADER+YN+PS VMSRLKDAGLNPD
Sbjct 38 RRQKRAIAAQKEENAQARAWSEKMARWYANNERANLADERSYNSPSTVMSRLKDAGLNPD 97
Query 98 LIYGNGAAGLVDSNVAGTapassvppadvagpIMATPTAMESLFQGAAYAKTLAETNNIK 157
LIYGNGAAGLVDSNVAGTAP SSVPPADVAGPIMATPTAMESLFQGAAYAKTLAET NIK
Sbjct 98 LIYGNGAAGLVDSNVAGTAPVSSVPPADVAGPIMATPTAMESLFQGAAYAKTLAETKNIK 157
Query 158 ADTSKKEGEVTSLNIDNFVKAA 179
ADTSKKEGEVTSLNIDNFVKAA
Sbjct 158 ADTSKKEGEVTSLNIDNFVKAA 179
> Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.protein.BACPLE
Length=383
Score = 47.8 bits (112), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 34/59 (58%), Gaps = 11/59 (19%)
Query 48 KEENAQARAWSEKMARWYANNERANLADERAYNNPSAVMSRLKDAGLNPDLIYGNGAAG 106
+EE + W E+M + NN +YN+P+A M RLK+AGLNPDL+Y G G
Sbjct 62 REERLAQQQWIEQM--YEKNN---------SYNSPAAQMQRLKEAGLNPDLMYSRGDVG 109
> Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.protein.BACPLE
Length=382
Score = 47.4 bits (111), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 34/59 (58%), Gaps = 11/59 (19%)
Query 48 KEENAQARAWSEKMARWYANNERANLADERAYNNPSAVMSRLKDAGLNPDLIYGNGAAG 106
+EE + W E+M + NN +YN+P+A M RLK+AGLNPDL+Y G G
Sbjct 61 REERLAQQQWIEQM--YEKNN---------SYNSPAAQMQRLKEAGLNPDLMYSRGDVG 108
> Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.protein.BACEGG.02723
Length=233
Score = 45.8 bits (107), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 25/56 (45%), Positives = 34/56 (61%), Gaps = 12/56 (21%)
Query 69 ERANLAD---------ERAYNNPSAVMSRLKDAGLNPDLIYGNGAAGLVDSNVAGT 115
+R NLA+ + YN P A M RLK+AGLNP+LIYG+G ++ VAG+
Sbjct 25 QRQNLANRQNVEFWNMQNKYNTPKAQMERLKEAGLNPNLIYGSGQ---TNTGVAGS 77
> Pichovirinae_66_Microbialites_001_Microviridae_AG0133_hypothetical.protein.BACEGG.02723
Length=275
Score = 44.3 bits (103), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 29/51 (57%), Gaps = 5/51 (10%)
Query 61 MARWYANNERANLAD-----ERAYNNPSAVMSRLKDAGLNPDLIYGNGAAG 106
+++ Y N ++ N A + YN P M R KDAGLNP LIYG G AG
Sbjct 23 LSQAYGNRQQRNFAVKFWNMQNEYNLPINQMQRFKDAGLNPHLIYGQGNAG 73
> Pichovirinae_Pavin_723_Microviridae_AG0326_hypothetical.protein.BACEGG.02723
Length=266
Score = 42.7 bits (99), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/55 (45%), Positives = 36/55 (65%), Gaps = 8/55 (15%)
Query 51 NAQARAWSEKMARWYANNERAN-LAD---ERAYNNPSAVMSRLKDAGLNPDLIYG 101
NAQ + +S++M + +RA+ L D + YN+PS M R K+AGLNP+LIYG
Sbjct 28 NAQNKKFSQEMY----DRQRADALQDWDKQNKYNSPSQQMQRYKEAGLNPNLIYG 78
> Pichovirinae_Bourget_523_Microviridae_AG0337_hypothetical.protein.BACEGG.02723
Length=307
Score = 41.6 bits (96), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 36/60 (60%), Gaps = 6/60 (10%)
Query 49 EENAQARAWSEKMARWYANNERANLA---DERAYNNPSAVMSRLKDAGLNPDLIYGNGAA 105
++N ++RA+S +M Y + NL + YN+P M+RLK AGLNP+++Y G A
Sbjct 28 KQNKKSRAFSREM---YEKTKADNLMFWDMQNQYNSPEQQMARLKSAGLNPNMVYDKGGA 84
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 39.3 bits (90), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 27/63 (43%), Positives = 32/63 (51%), Gaps = 13/63 (21%)
Query 53 QARAWSEKMARWYANNERANLADERAYNNPSAVMSRLKDAGLNPDLIYGNGAAGLVDSNV 112
Q R W E M W E AYN SA RL++AGLNP L+ G+AG+ S
Sbjct 52 QQRNWQENM--W---------NKENAYNTASAQRQRLEEAGLNPYLMMNGGSAGVAQS-- 98
Query 113 AGT 115
AGT
Sbjct 99 AGT 101
> Pichovirinae_JCVI_003_Microviridae_AG0346_hypothetical.protein.BACEGG.02723
Length=252
Score = 38.1 bits (87), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/55 (42%), Positives = 31/55 (56%), Gaps = 6/55 (11%)
Query 53 QARAWSEKMARWYANNERANLAD-ERA--YNNPSAVMSRLKDAGLNPDLIYGNGA 104
+ RA+ E++ Y + L D +R YN+P M RLK+AGLNP L YG A
Sbjct 32 EQRAYEEQI---YERQRKDALEDWQRTTDYNSPEQQMQRLKEAGLNPKLAYGTLA 83
> Pichovirinae_Pavin_279_Microviridae_AG0269_hypothetical.protein.BACPLE
Length=292
Score = 37.4 bits (85), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 25/37 (68%), Gaps = 2/37 (5%)
Query 74 ADERAYNNPSAVMSRLKDAGLNPDLIYGNGAAGLVDS 110
A + YN+P+A RLKDA LNP L+Y G +G V+S
Sbjct 68 AAQNVYNSPAAQKQRLKDAELNPSLMY--GGSGHVES 102
Lambda K H a alpha
0.311 0.123 0.340 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 13123488