bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
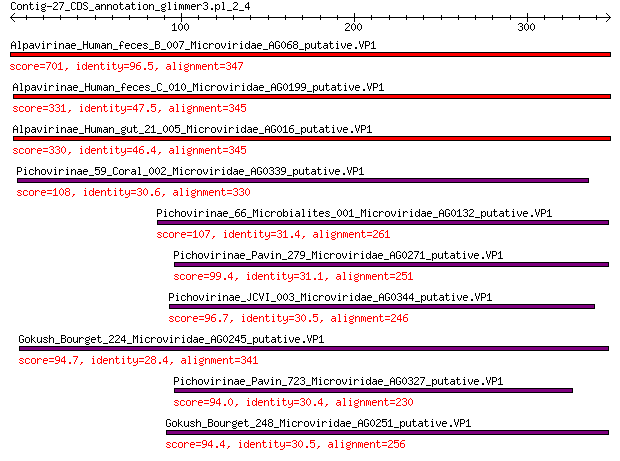
Query= Contig-27_CDS_annotation_glimmer3.pl_2_4
Length=347
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 701 0.0
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 331 2e-108
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 330 6e-108
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 108 7e-28
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 107 1e-27
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 99.4 8e-25
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 96.7 6e-24
Gokush_Bourget_224_Microviridae_AG0245_putative.VP1 94.7 3e-23
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 94.0 5e-23
Gokush_Bourget_248_Microviridae_AG0251_putative.VP1 94.4 5e-23
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 701 bits (1810), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 335/347 (97%), Positives = 343/347 (99%), Gaps = 0/347 (0%)
Query 1 LYANWQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVD 60
LYANWQSDAYTTAL APQQ V PLVGLTTYE+VS+N+AGHTVTTVNTAIVDE+GNAYKVD
Sbjct 434 LYANWQSDAYTTALTAPQQGVAPLVGLTTYESVSLNDAGHTVTTVNTAIVDEDGNAYKVD 493
Query 61 FESNGDALKGVNYTPLKAGEAVNMQSLVSPVASGISINDFRNVNAYQRYLELNQFRGFSY 120
FESNG+ALKGVNYTPLKAGEA+NMQSLVSPV SGISINDFRNVNAYQRYLELNQFRGFSY
Sbjct 494 FESNGEALKGVNYTPLKAGEAINMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSY 553
Query 121 KEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQAGLATC 180
KEIIEGRFDVNVRYDALNMPEYLGGITRDI+VNPITQTVETTD+GSYVGSLGSQAGLATC
Sbjct 554 KEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETTDTGSYVGSLGSQAGLATC 613
Query 181 FGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIY 240
FGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIY
Sbjct 614 FGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIY 673
Query 241 LKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELG 300
LKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELG
Sbjct 674 LKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELG 733
Query 301 KSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 347
KSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE
Sbjct 734 KSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 780
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 331 bits (849), Expect = 2e-108, Method: Compositional matrix adjust.
Identities = 164/360 (46%), Positives = 235/360 (65%), Gaps = 15/360 (4%)
Query 3 ANWQSDAYTTALAAPQQDV-VPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDF 61
NW+ D TTA+ PQQ PLVGL + V+ +E G T +VDE+G+ Y V +
Sbjct 372 CNWERDFLTTAVPNPQQGANAPLVGLVMGDVVTRSEDGTYSVQKQTVLVDEDGSKYGVSY 431
Query 62 E--SNGDALKGVNYTPLKA-------GEAVNMQSLVSPVASGISINDFRNVNAYQRYLEL 112
+ +G+ L GV+Y P+ G + +L + SG +I R VNAYQ++LEL
Sbjct 432 KVSEDGERLVGVDYDPVSEKTPVTAIGSYAELAALATEQGSGFTIETLRYVNAYQKFLEL 491
Query 113 NQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVET---TDSGSYVG 169
N +GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV+ + G Y
Sbjct 492 NMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSGSSRGQYAE 551
Query 170 SLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNP 229
+LGS+ G+A +G++ +I VFCDEES ++G++ V P+P+Y +LPK TY LD + P
Sbjct 552 ALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQP 611
Query 230 EFDHIGYQPIYLKELAA--IQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNF 287
EFD IG+QPI KE+ I A +S LN FGYQRPWYEYV K D AHGLF ++++NF
Sbjct 612 EFDRIGFQPITYKEICPMNIVAGDSANQLNKTFGYQRPWYEYVAKYDSAHGLFRTNMKNF 671
Query 288 IMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 347
IM R F P+LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 672 IMSRVFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 330 bits (846), Expect = 6e-108, Method: Compositional matrix adjust.
Identities = 160/359 (45%), Positives = 234/359 (65%), Gaps = 14/359 (4%)
Query 3 ANWQSDAYTTALAAPQQDV-VPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDF 61
NW+ D TTA+ PQQ PLVGLT + V+ +E G T +VDE+G+ Y + +
Sbjct 384 CNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRSEDGTLSVQKQTVLVDEDGSKYGISY 443
Query 62 E--SNGDALKGVNYTPLKAGEAVN-------MQSLVSPVASGISINDFRNVNAYQRYLEL 112
+ +G+ L GV+Y P+ V + +L + SG +I R VNAYQ++LEL
Sbjct 444 KVSEDGERLVGVDYDPVSEKTPVTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLEL 503
Query 113 NQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVE---TTDSGSYVG 169
N +GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV+ + G Y
Sbjct 504 NMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSASSQGQYAE 563
Query 170 SLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNP 229
+LGS+ G+A +G++ +I VFCDEES ++G++ V P+P+Y +LPK TY LD + P
Sbjct 564 ALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYSQLLPKDFTYNGLLDHYQP 623
Query 230 EFDHIGYQPIYLKELAAIQ-AFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFI 288
EFD IG+QPI KE+ + + D+N FGYQRPWYEYV K D AHGLF + L+ F+
Sbjct 624 EFDRIGFQPITYKEVCPLNFDISNMNDMNKTFGYQRPWYEYVAKYDNAHGLFRTDLKKFV 683
Query 289 MFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 347
M R+F P+LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 684 MHRTFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 742
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 108 bits (269), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 101/354 (29%), Positives = 147/354 (42%), Gaps = 50/354 (14%)
Query 5 WQSDAYTTALAAPQQD---VVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDF 61
WQ D +T+AL Q+ +PL G + T S N A T+ NT+
Sbjct 183 WQHDYFTSALPWTQRGPEATIPL-GTSAPITWSNNTATATIVRNNTS------------- 228
Query 62 ESNGDALKGVNYTPLKA----GEAVNMQSLVSPV----------------ASGISINDFR 101
G+ + G + A G + L +PV A+ SIND R
Sbjct 229 ---GNPIAGYTFDGASALYTGGSGQLIADLPTPVSLDFDNSDHLFADLAGATASSINDLR 285
Query 102 NVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVET 161
Q +LE N G Y EII F V L PE+LGG I I++ ++T
Sbjct 286 RAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPIT---ISEVLQT 342
Query 162 TDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYR 221
+ + + G+ AG G+S+ +S C+E ++GIM VMP Y +PK
Sbjct 343 SANNTEPTPQGNMAGHGVSVGSSN-YVSYKCEEHGYIIGIMSVMPKTAYQQGIPKHFKKL 401
Query 222 ERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNT-VFGYQRPWYEYVQKVDRAHGLF 280
++ D + P F +IG QPI +EL + +T FGY + EY HG F
Sbjct 402 DKFDYYWPSFANIGEQPILNEEL-----YHQNNATDTETFGYTPRYAEYKYIPSTVHGEF 456
Query 281 LSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTA 334
SL + M R F + P L + F V VF+VT + + +H + A
Sbjct 457 RDSLDFWHMGRIFGSKPTLNQDFIECNADDVERVFAVTAGQEHLYVYLHNEVKA 510
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 107 bits (267), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 82/263 (31%), Positives = 123/263 (47%), Gaps = 13/263 (5%)
Query 86 SLVSPV-ASGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLG 144
SLV V A + IN R Q +LE N G Y E I F V L PEYLG
Sbjct 267 SLVVDVQADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLG 326
Query 145 GITRDIIVNPITQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYV 204
G +++++ + T ETTD+ VG++ G+ GN +I C+E ++GI+ V
Sbjct 327 GTKGNMVISEVLATAETTDANVAVGTMAGH-GITAHGGN---TIKYRCEEHGWIIGIINV 382
Query 205 MPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQR 264
P+ Y LP+ + +RLD P F +IG Q +Y KEL A + + +FGY
Sbjct 383 QPVTAYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKELYA-----NSPAPDEIFGYIP 437
Query 265 PWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTE-VSDK 323
+ E K R G +L + + R F N P L + F P + +F+VT+ D
Sbjct 438 RYAEMKFKNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECNPDT--RIFAVTDPAEDH 495
Query 324 ILGQIHFDCTAQLPISRVVVPRL 346
I+ I+ + T + R +P +
Sbjct 496 IVAHIYNEVTVNRKLPRYGIPTI 518
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 99.4 bits (246), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 78/253 (31%), Positives = 115/253 (45%), Gaps = 14/253 (6%)
Query 96 SINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPI 155
+IND R Q +LE N G Y E I+ F V L PEY+ G ++V I
Sbjct 263 TINDLRRAMRLQEFLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEI 322
Query 156 TQTVETTDSGSYVGSLGSQAGLATCFGNSD-GSISVFCDEESIVMGIMYVMPMPVYDSIL 214
T TV+ + G+ G AT + G+ SV +E ++GIM +MP P Y +
Sbjct 323 TSTVKGIEEPQ-----GNPTGKATSLQSGHYGNYSV--EEHGYIIGIMSIMPKPAYQQGI 375
Query 215 PKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVD 274
PK + LD + P F +IG Q + +E+ + N VFGY + EY +
Sbjct 376 PKSYLKTDVLDFYWPSFANIGEQEVLQEEI-----YGYTPTPNAVFGYIPRYAEYKYLQN 430
Query 275 RAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTE-VSDKILGQIHFDCT 333
R GLF ++L + + R F N P L +SF + P + +F+ E D I + T
Sbjct 431 RVCGLFRTTLDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNIYIHVLNKMT 490
Query 334 AQLPISRVVVPRL 346
A+ P+ P L
Sbjct 491 ARRPMPVFGTPML 503
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 96.7 bits (239), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 75/248 (30%), Positives = 113/248 (46%), Gaps = 12/248 (5%)
Query 93 SGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIV 152
+ +I D R Q +LEL G Y E I+ FDV L PEY+ G + +I+
Sbjct 262 TSTTITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVII 321
Query 153 NPITQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDS 212
+ I T T +G G++ S G+A GN +C+E ++GI+ V P Y
Sbjct 322 SEILNTTGET-AGLPQGNM-SGHGVAVGTGNVG---KYYCEEHGFIIGILSVTPNTAYQQ 376
Query 213 ILPKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQK 272
+PK ++ D + P+F HIG QPI E I A++ D FGY + EY
Sbjct 377 GIPKHYLRTDKYDFYWPQFAHIGEQPIVNNE---IYAYDPTGD--ETFGYTPRYAEYKYM 431
Query 273 VDRAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDC 332
R G F SSL + R F + P L + F + + +F+VT+ D + +
Sbjct 432 PSRVAGEFRSSLDEWHAARIFSSLPTLSQDFIEVDADDFDRIFAVTDGDDNLYMHVLNKV 491
Query 333 TA--QLPI 338
A Q+P+
Sbjct 492 KARRQMPV 499
> Gokush_Bourget_224_Microviridae_AG0245_putative.VP1
Length=536
Score = 94.7 bits (234), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 97/350 (28%), Positives = 145/350 (41%), Gaps = 21/350 (6%)
Query 6 QSDAYTTALAAPQQDVVPL-VGLTTYETVSVNEAGHTVTTVNT--AIVDEEGNAYKVDFE 62
+ D +T AL PQ+ P+ + + T+ V N+ + + N A+ +++ A + +
Sbjct 195 RHDYFTGALPWPQKGGTPVTIPIGTFAPVLWNKYAGSPSNFNNTNAVFEQQVTAADLRYA 254
Query 63 SNGDALKGVNYTPLKAGEAVNMQSLVSPVASGISINDFRNVNAYQRYLELNQFRGFSYKE 122
G T L G+ + + +S A+ ++IN R Q+ LE + G Y E
Sbjct 255 PT----YGGASTALAPGDVGGLYTDLS-NATAVTINQLRQSFQIQKLLERDARGGTRYTE 309
Query 123 IIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQAGLATCFG 182
I+ F V L PEYLGG T I ++PI QT T SG G+ A + T
Sbjct 310 ILRSHFGVTSPDARLQRPEYLGGGTTPISISPIAQTSGTGTSGQTT-PQGNLAAMGTYLA 368
Query 183 NSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIYLK 242
G F E V+G++ V Y L + + R D + P F +G Q I K
Sbjct 369 KGHGFSQSFV-EHGYVIGVVSVRADLTYQQGLRRHWSRSTRYDYYFPAFAMLGEQSILNK 427
Query 243 ELAAIQAFESGKDL-NTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFI----MFRSFENAP 297
E+ + +G L N VFGYQ W EY GLF S+ I + F + P
Sbjct 428 EI-----YVTGGSLDNDVFGYQERWAEYRYNPSEITGLFRSTAAGTIDPWHYAQKFTSLP 482
Query 298 ELGKSFTVMQPGSVNNV-FSVTEVSDKILGQIHFDCTAQLPISRVVVPRL 346
L +F P N+ + + L FD A P+ VP L
Sbjct 483 TLNTTFIQDNPPLARNLAVGASANGQQFLLDAFFDINAARPLPMYSVPGL 532
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 94.0 bits (232), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 70/230 (30%), Positives = 101/230 (44%), Gaps = 12/230 (5%)
Query 96 SINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPI 155
+IND R Q +LE N G Y E I F V L PEY+ G+ ++V+ +
Sbjct 268 TINDLRRAFKLQEWLEKNARGGTRYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEV 327
Query 156 TQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILP 215
T D G G++ G++ G S S +C+E ++GIM VMP Y +P
Sbjct 328 LNTT-GQDGGLPQGNMAGH-GISVTSGKSG---SYYCEEHGYIIGIMSVMPKTAYQQGIP 382
Query 216 KWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDR 275
+ + LD F P F +IG Q + +EL A A + N FGY + EY R
Sbjct 383 RTFLKTDSLDYFWPTFANIGEQEVAKQELYAYTA-----NANDTFGYVPRYAEYKYMPSR 437
Query 276 AHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKIL 325
G F +SL + + R F P L F P +F+V + +L
Sbjct 438 VAGEFRTSLNYWHLGRIFATEPSLNSDFIECDP--TKRIFAVEDPETDVL 485
> Gokush_Bourget_248_Microviridae_AG0251_putative.VP1
Length=546
Score = 94.4 bits (233), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 78/262 (30%), Positives = 115/262 (44%), Gaps = 13/262 (5%)
Query 91 VASGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDI 150
VA+ +IN R Q+ LE + G Y EII F V L PEY+GG + +I
Sbjct 288 VATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVISPDARLQRPEYIGGGSTNI 347
Query 151 IVNPITQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVY 210
+NPI QT T SG+ +G+ A + T ++ G + E +++G++ V Y
Sbjct 348 NINPIAQTSGTNASGTTT-PMGTLAAMGTALAHNHG-FTYSSTEHGVIIGMVSVRADLTY 405
Query 211 DSILPKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYV 270
L + + R D + P F +G Q + KE I + D N VFGYQ W EY
Sbjct 406 QQGLQRMWSRSTRYDFYFPAFATLGEQAVLNKE---IYVTGNAGD-NDVFGYQERWAEYR 461
Query 271 QKVDRAHGLFLSSLRNFI----MFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSD--KI 324
R LF S+ I + + F P L +F P V+ V +V ++ +
Sbjct 462 YYPSRISSLFRSTASGTIDAWHLAQKFTTTPTLNNTFIQDTP-PVSRVVAVGAAANGQQF 520
Query 325 LGQIHFDCTAQLPISRVVVPRL 346
+ FDC P+ VP L
Sbjct 521 IFDSFFDCKKARPMPMYSVPGL 542
Lambda K H a alpha
0.318 0.135 0.396 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 29958948