bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
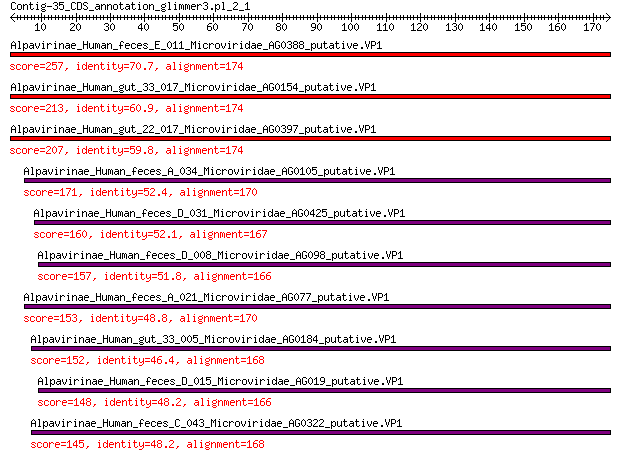
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 257 7e-84
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 213 3e-67
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 207 5e-65
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 171 9e-52
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 160 7e-48
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 157 8e-47
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 153 2e-45
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 152 7e-45
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 148 1e-43
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 145 8e-43
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 257 bits (656), Expect = 7e-84, Method: Compositional matrix adjust.
Identities = 123/175 (70%), Positives = 139/175 (79%), Gaps = 2/175 (1%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
M Y+TGS+YCIIMCIYHA+PLLDYAI+GQD QLL T++EDLPIPEFDNIGMEAVP L
Sbjct 449 MRYHTGSQYCIIMCIYHAMPLLDYAITGQDPQLLCTSVEDLPIPEFDNIGMEAVPATTLF 508
Query 61 NSSLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTSS 120
NS L F GYNPRY+ WK+KIDRVHGAFTT LKDWVAPIDD YL W
Sbjct 509 NSVLFD-GTAVNDFLGYNPRYWPWKSKIDRVHGAFTTTLKDWVAPIDDDYLHNWFNSKDG 567
Query 121 Q-PSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
+ S+ +PFFKVNPNTLD+IFAV ADS W TDQLL+NC++ CKVVRPLSQDG+PY
Sbjct 568 KSASISWPFFKVNPNTLDSIFAVVADSIWETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 213 bits (542), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 106/182 (58%), Positives = 133/182 (73%), Gaps = 13/182 (7%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
TY T +++C+IM IYHA+PLLDY ++GQDGQLLVT+ E LPIPEFDNIG+E +P+ Q+
Sbjct 438 FTYTT-NEHCVIMGIYHAVPLLDYTLTGQDGQLLVTDAESLPIPEFDNIGLEVLPMAQIF 496
Query 61 NSSLLSTNIGKYSFF--GYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPL- 117
NSSL + ++ F GYNPRY+NWKTK+D ++GAFTT LK WV+P+ +S L W
Sbjct 497 NSSLATA----FNLFNAGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVSESLLSGWARFG 552
Query 118 -----TSSQPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGI 172
T ++ L Y FFKVNP+ LD IF VKADSTW TDQLLVN I C VVR LS+DG+
Sbjct 553 ASDSKTGTKAVLNYKFFKVNPSVLDPIFGVKADSTWDTDQLLVNSYIGCYVVRNLSRDGV 612
Query 173 PY 174
PY
Sbjct 613 PY 614
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 207 bits (526), Expect = 5e-65, Method: Compositional matrix adjust.
Identities = 104/182 (57%), Positives = 127/182 (70%), Gaps = 11/182 (6%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
TY T +++C++MCIYHA+PLLDY I+GQDGQLLVT+ E LPIPEFDNIGME +P+ Q+
Sbjct 429 FTYTT-NEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMETLPMTQIF 487
Query 61 NSSLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTSS 120
NS S I GYNPRY+NWKTK+D ++GAFTT LK WV+P+ +S L W S
Sbjct 488 NSPKAS--IVNLFNAGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVTESLLSGWFGFGYS 545
Query 121 QPS--------LLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGI 172
+ L Y FFKVNP+ LD IF V ADSTW +DQLLVN I C V R LS+DG+
Sbjct 546 EGDVNSQNKVVLNYKFFKVNPSVLDPIFGVAADSTWDSDQLLVNSYIGCYVARNLSRDGV 605
Query 173 PY 174
PY
Sbjct 606 PY 607
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 171 bits (433), Expect = 9e-52, Method: Compositional matrix adjust.
Identities = 89/175 (51%), Positives = 116/175 (66%), Gaps = 7/175 (4%)
Query 5 TGSKYCIIMCIYHAIPLLDYAISGQDGQLLVT-NIEDLPIPEFDNIGMEAVPVVQLVNSS 63
T +++ +IMCIYHA PLLDY +G D QL+ T E P+PEFD++GME++P++ LVNS
Sbjct 446 TCNEHGVIMCIYHATPLLDYVPTGPDLQLMSTVKGESWPVPEFDSLGMESLPMLSLVNSK 505
Query 64 LLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQ----QWLPLTS 119
+ + + S+ GY PRY +WKT ID V GAFT LK W AP+D Y+ + +P
Sbjct 506 AIGDIVAR-SYAGYVPRYISWKTSIDVVRGAFTDTLKSWTAPVDSDYMHVFFGEVIPQEG 564
Query 120 SQPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
S P L Y +FKVNP+ L+ IFAV D +W TDQLL NC D KV R LS DG+PY
Sbjct 565 S-PILSYTWFKVNPSVLNPIFAVSVDGSWNTDQLLCNCQFDVKVARNLSYDGMPY 618
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 160 bits (405), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 87/175 (50%), Positives = 111/175 (63%), Gaps = 8/175 (5%)
Query 8 KYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSLLST 67
+Y II+ IYH +PLLDY I+GQ LL TN DLP PEFD+IGM+ + + VNS +S
Sbjct 440 EYGIIIGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEFDSIGMQTIQFGRFVNSKSVSW 499
Query 68 NIG---KYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWL--PLTSSQP 122
G + GY PR+++ KT+ D V GAF + LK+WVAP+D SY+ +WL +TSS
Sbjct 500 TSGVDYRVQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPSYVSKWLQSSVTSSGK 559
Query 123 ---SLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
+L Y FFKVNP LD+IF VK DST TDQ L +D K VR DG+PY
Sbjct 560 LALNLNYGFFKVNPRVLDSIFNVKCDSTIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 157 bits (397), Expect = 8e-47, Method: Compositional matrix adjust.
Identities = 86/174 (49%), Positives = 107/174 (61%), Gaps = 8/174 (5%)
Query 9 YCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSLLSTN 68
Y IIM IYH +PLLDY I+GQ LL TN DLP PE+D+IGM+ + + VNS +
Sbjct 445 YGIIMGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEYDSIGMQTIQFGRFVNSKAVGWT 504
Query 69 IG---KYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWL--PLTSSQPS 123
G + GY PR+++ KT+ D V GAF + LK+WVAP+D + L QWL +TSS
Sbjct 505 SGVDYRTQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPANLPQWLQTSVTSSGKL 564
Query 124 LL---YPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
L Y FFKVNP LD+IF VK DST TDQ L +D K VR DG+PY
Sbjct 565 FLNLNYGFFKVNPRVLDSIFNVKCDSTIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 153 bits (386), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 83/180 (46%), Positives = 107/180 (59%), Gaps = 10/180 (6%)
Query 5 TGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSL 64
T ++ IIM IYHA+PLLDY +GQD LL+T+ E+ IPEFD IGM+ +P+ L NS+
Sbjct 443 TARQHGIIMGIYHAVPLLDYERTGQDQDLLITSAEEWAIPEFDAIGMQTLPLGTLFNSNK 502
Query 65 LSTNI-----GKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTS 119
+S + G GY PRY NWKT ID + GAF + K WVAPID ++ W+ +
Sbjct 503 VSGDSDFRLHGAAYPIGYVPRYVNWKTDIDEIFGAFRSSEKTWVAPIDADFITNWVKNVA 562
Query 120 SQPSLL-----YPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
S + Y +FKVNP LD+IFAVKADST TD + K VR L G+PY
Sbjct 563 DNASAVQSLFNYNWFKVNPAILDDIFAVKADSTMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 152 bits (383), Expect = 7e-45, Method: Compositional matrix adjust.
Identities = 78/172 (45%), Positives = 109/172 (63%), Gaps = 5/172 (3%)
Query 7 SKYCIIMCIYHAIPLLDYAISGQDGQLLVT-NIEDLPIPEFDNIGMEAVPVVQLVNSSLL 65
+++ ++MCIYHA+PLLDYA +G D Q + T + + P+PE D+IG E +P L+N++ +
Sbjct 448 TEHGVLMCIYHAVPLLDYAPTGPDLQFMTTVDGDSWPVPEMDSIGFEELPSYSLLNTNAV 507
Query 66 STNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLP---LTSSQP 122
I + FGY PRY +WKT +D V GAF LK W API YL+ + +
Sbjct 508 QP-IKEPRPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGQDYLKIYFDNNNVPGGAH 566
Query 123 SLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
Y +FKVNP+ ++ IF V AD +W TDQLLVNC+ D +V R LS DG+PY
Sbjct 567 FGFYTWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 618
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 148 bits (374), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 80/172 (47%), Positives = 100/172 (58%), Gaps = 6/172 (3%)
Query 9 YCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSLLSTN 68
+ ++MCIYH++PLLDY IS D QL + P+PE D IG+E + V N+ + +
Sbjct 413 WGVLMCIYHSVPLLDYVISAPDPQLFASMNTSFPVPELDAIGLEPITVAYYSNNPIELPS 472
Query 69 IGKYS-----FFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWL-PLTSSQP 122
G + GY PRYY WKT ID V GAFTT K+WVAPI L PL +
Sbjct 473 TGGITDAPTTTVGYLPRYYAWKTSIDYVLGAFTTTEKEWVAPITPELWSNMLKPLGTKGT 532
Query 123 SLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
+ Y FFKVNP+ LD IFAV ADS W TD L+N D +V R L DG+PY
Sbjct 533 GINYNFFKVNPSILDPIFAVNADSYWDTDTFLINAAFDIRVARNLDYDGMPY 584
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 145 bits (367), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 81/173 (47%), Positives = 108/173 (62%), Gaps = 5/173 (3%)
Query 7 SKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVN--SSL 64
+++ I+MCIYHA P+LDY SGQD QLL T D+PIPEFD+IGMEA+P+ L N S+
Sbjct 430 NEHGILMCIYHATPVLDYLRSGQDLQLLSTLATDIPIPEFDHIGMEALPIETLFNEQSTE 489
Query 65 LSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPID-DSYLQQWL--PLTSSQ 121
+ I GY+PRY +KT +D V GAF T L WVAP+ + + + L P + S
Sbjct 490 ATALINSIPVLGYSPRYIAYKTSVDWVSGAFETTLDSWVAPLTVNEQITKLLFNPDSGSV 549
Query 122 PSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
S+ Y FFKV P LD IF + TW +DQ LVN + + K V+ L +G+PY
Sbjct 550 YSMNYGFFKVTPRVLDPIFVQECTDTWDSDQFLVNVSFNVKPVQNLDYNGMPY 602
Lambda K H a alpha
0.321 0.139 0.440 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 12344976