bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-38_CDS_annotation_glimmer3.pl_2_1
Length=229
Score E
Sequences producing significant alignments: (Bits) Value
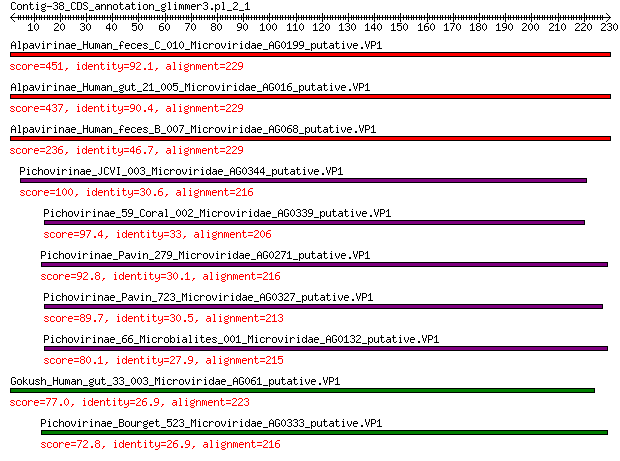
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 451 1e-156
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 437 2e-151
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 236 8e-74
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 100 4e-26
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 97.4 6e-25
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 92.8 2e-23
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 89.7 3e-22
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 80.1 5e-19
Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1 77.0 5e-18
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 72.8 1e-16
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 451 bits (1159), Expect = 1e-156, Method: Compositional matrix adjust.
Identities = 211/229 (92%), Positives = 221/229 (97%), Gaps = 0/229 (0%)
Query 1 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGV 60
MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQS SS+GQYAEALGSK+GIAGV
Sbjct 503 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSGSSRGQYAEALGSKTGIAGV 562
Query 61 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPIT 120
YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQ+L KDF YNGLLDHYQPEFDRIGFQPIT
Sbjct 563 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPIT 622
Query 121 YKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQ 180
YKE+CP+N+ D+ N+ N+TFGYQRPWYEYVAKYDSAHGLFRTNMKNF+MSRVFSGLPQ
Sbjct 623 YKEICPMNIVAGDSANQLNKTFGYQRPWYEYVAKYDSAHGLFRTNMKNFIMSRVFSGLPQ 682
Query 181 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 229
LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 683 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 437 bits (1125), Expect = 2e-151, Method: Compositional matrix adjust.
Identities = 207/229 (90%), Positives = 218/229 (95%), Gaps = 1/229 (0%)
Query 1 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGV 60
MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQS SSQGQYAEALGSK+GIAGV
Sbjct 515 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSASSQGQYAEALGSKTGIAGV 574
Query 61 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPIT 120
YGSTSNNIEVFCDEESYIIGLLTVTPVPIY+Q+L KDF YNGLLDHYQPEFDRIGFQPIT
Sbjct 575 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYSQLLPKDFTYNGLLDHYQPEFDRIGFQPIT 634
Query 121 YKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQ 180
YKEVCPLN +++ +N N+TFGYQRPWYEYVAKYD+AHGLFRT++K FVM R FSGLPQ
Sbjct 635 YKEVCPLNFDISN-MNDMNKTFGYQRPWYEYVAKYDNAHGLFRTDLKKFVMHRTFSGLPQ 693
Query 181 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 229
LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 694 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 236 bits (601), Expect = 8e-74, Method: Compositional matrix adjust.
Identities = 107/229 (47%), Positives = 160/229 (70%), Gaps = 5/229 (2%)
Query 1 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGV 60
++GR+D+++R+D L MPE++GGI+R++ + + QTV+ T+ G Y +LGS++G+A
Sbjct 557 IEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVE---TTDTGSYVGSLGSQAGLATC 613
Query 61 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPIT 120
+G++ +I VFCDEES ++G++ V P+P+Y +L K Y LD + PEFD IG+QPI
Sbjct 614 FGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIY 673
Query 121 YKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQ 180
KE+ + ++ N FGYQRPWYEYV K D AHGLF ++++NF+M R F P+
Sbjct 674 LKELAAIQ--AFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPE 731
Query 181 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 229
LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 732 LGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 780
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 100 bits (249), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 66/218 (30%), Positives = 108/218 (50%), Gaps = 17/218 (8%)
Query 5 WDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGST 64
+D+ L PE+I G + + + + T + + QG + SG GV T
Sbjct 297 FDVKSSDQRLQRPEYITGTKQPVIISEILNTTGETAGLPQG-------NMSG-HGVAVGT 348
Query 65 SNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEV 124
N + +C+E +IIG+L+VTP Y Q + K +L D Y P+F IG QPI E+
Sbjct 349 GNVGKYYCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEI 408
Query 125 CPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQ 184
+ ++TFGY + EY G FR+++ + +R+FS LP L Q
Sbjct 409 YAYD-------PTGDETFGYTPRYAEYKYMPSRVAGEFRSSLDEWHAARIFSSLPTLSQD 461
Query 185 FLLVDPDTVNQVFSVTEYTDKIFGYV--KFNATARLPI 220
F+ VD D +++F+VT+ D ++ +V K A ++P+
Sbjct 462 FIEVDADDFDRIFAVTDGDDNLYMHVLNKVKARRQMPV 499
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 97.4 bits (241), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 68/209 (33%), Positives = 102/209 (49%), Gaps = 17/209 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQT-VDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PEF+GG + +++ V QT + + QG A GV +SN + C
Sbjct 321 LQRPEFLGGSATPITISEVLQTSANNTEPTPQGNMAGH--------GVSVGSSNYVSYKC 372
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E YIIG+++V P Y Q + K F D+Y P F IG QPI +E+ N
Sbjct 373 EEHGYIIGIMSVMPKTAYQQGIPKHFKKLDKFDYYWPSFANIGEQPILNEELYHQN-NAT 431
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
DT +TFGY + EY + HG FR ++ + M R+F P L Q F+ + D
Sbjct 432 DT-----ETFGYTPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFGSKPTLNQDFIECNADD 486
Query 193 VNQVFSVTEYTDKIFGYV--KFNATARLP 219
V +VF+VT + ++ Y+ + AT +P
Sbjct 487 VERVFAVTAGQEHLYVYLHNEVKATRLMP 515
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 92.8 bits (229), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 65/217 (30%), Positives = 98/217 (45%), Gaps = 17/217 (8%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PE+I G + ++ + TV +A +SG G Y
Sbjct 303 RLQRPEYITGTKAPVVVQEITSTVKGIEEPQGNPTGKATSLQSGHYGNYS---------V 353
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E YIIG++++ P P Y Q + K +L +LD Y P F IG Q + +E+ G
Sbjct 354 EEHGYIIGIMSIMPKPAYQQGIPKSYLKTDVLDFYWPSFANIGEQEVLQEEI----YGYT 409
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
T N FGY + EY + GLFRT + + + R+F+ +P L Q F+ V P
Sbjct 410 PT---PNAVFGYIPRYAEYKYLQNRVCGLFRTTLDFWHLGRIFANIPTLSQSFIEVSPAN 466
Query 193 VNQVFSVTE-YTDKIFGYVKFNATARLPISRVAIPRL 228
++F+ E Y D I+ +V TAR P+ P L
Sbjct 467 QTRIFANQEDYDDNIYIHVLNKMTARRPMPVFGTPML 503
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 89.7 bits (221), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 65/215 (30%), Positives = 100/215 (47%), Gaps = 20/215 (9%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYA-EALGSKSGIAGVYGSTSNNIEVFC 72
L PE+I G+ + + V T Q QG A + SG +G Y +C
Sbjct 309 LQRPEYITGVKSPVVVSEVLNTTGQDGGLPQGNMAGHGISVTSGKSGSY---------YC 359
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E YIIG+++V P Y Q + + FL LD++ P F IG Q + +E+ A
Sbjct 360 EEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANIGEQEVAKQEL------YA 413
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
T N AN TFGY + EY G FRT++ + + R+F+ P L F+ DP
Sbjct 414 YTAN-ANDTFGYVPRYAEYKYMPSRVAGEFRTSLNYWHLGRIFATEPSLNSDFIECDP-- 470
Query 193 VNQVFSVTE-YTDKIFGYVKFNATARLPISRVAIP 226
++F+V + TD ++ +V A P+ + P
Sbjct 471 TKRIFAVEDPETDVLYCHVLNKIKAVRPMPKYGTP 505
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 80.1 bits (196), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 60/216 (28%), Positives = 99/216 (46%), Gaps = 17/216 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCD 73
L PE++GG + + V T + + A+G+ +G G+ N I+ C+
Sbjct 319 LQRPEYLGGTKGNMVISEVLATAETTDANV------AVGTMAG-HGITAHGGNTIKYRCE 371
Query 74 EESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVAD 133
E +IIG++ V PV Y Q L ++F LD+ P F IG Q + KE+ N D
Sbjct 372 EHGWIIGIINVQPVTAYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKELY-ANSPAPD 430
Query 134 TVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTV 193
+ FGY + E K G R + + + R+F+ P L ++F+ +PDT
Sbjct 431 EI------FGYIPRYAEMKFKNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECNPDT- 483
Query 194 NQVFSVTE-YTDKIFGYVKFNATARLPISRVAIPRL 228
++F+VT+ D I ++ T + R IP +
Sbjct 484 -RIFAVTDPAEDHIVAHIYNEVTVNRKLPRYGIPTI 518
> Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1
Length=545
Score = 77.0 bits (188), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 60/232 (26%), Positives = 104/232 (45%), Gaps = 25/232 (11%)
Query 1 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGV 60
+Q WD+ I + +PE++GG ++M + QT QQ+ + G G
Sbjct 324 VQALWDVVISDKTMQIPEYLGGGRYHVNMNQIVQTSGQQTNDD---------TPIGETGA 374
Query 61 YGSTSNNIEVFC---DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQ 117
T N F +E ++IG+ V Y Q L + + LD+Y P+F +G Q
Sbjct 375 MSVTPINESSFTKSFEEHGFVIGVCCVRHNHSYQQGLERFWSRTDRLDYYVPQFANLGEQ 434
Query 118 PITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKN----FVMSR 173
PI KE+ + G A +TFGYQ W +Y K + GL R+N + + +
Sbjct 435 PIKKKEI--MLTGTASD----EETFGYQEAWADYRMKPNRVSGLMRSNAEGTLDFWHYAD 488
Query 174 VFSGLPQLGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKF--NATARLPISRV 223
+S +P L Q+++ + + + + + + FG ++ T R+P+ V
Sbjct 489 NYSTVPTLSQEWIAEGKEEIARTL-IVQNEPQFFGAIRIANKTTRRMPLYSV 539
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 72.8 bits (177), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 58/217 (27%), Positives = 98/217 (45%), Gaps = 20/217 (9%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PE++GG ++ + V T + +G+ +G G+ S N +
Sbjct 321 RLQRPEYLGGSKGKMVISEVLSTAET---------TLPVGNMAG-HGISVSGGNEFKYNV 370
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E +IIG+++VTP Y Q + + LD++ P F IG Q + KE+ A
Sbjct 371 EEHGWIIGIISVTPETAYQQGIHRSLSKLDRLDYFWPTFANIGEQEVLGKEI------YA 424
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
+ +N +TFGY + EY G RT++ + + R FS P L F+ DP T
Sbjct 425 NGIN-IGETFGYVPRYAEYKFLNSRVAGEMRTSLDYWHLGRKFSAAPNLNGAFIECDPST 483
Query 193 VNQVFSVTEY-TDKIFGYVKFNATARLPISRVAIPRL 228
++F+V + D I+G++ N A + + P
Sbjct 484 --RIFAVEDAEVDNIYGHIFNNIKAIRKMPKYGTPNF 518
Lambda K H a alpha
0.321 0.138 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 17847033