bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_6
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
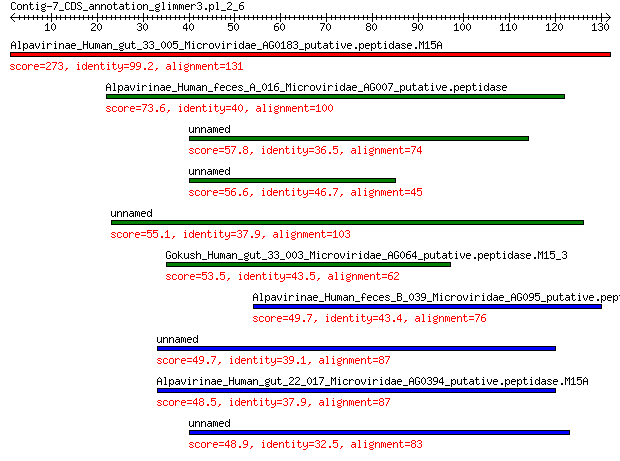
Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.pepti... 273 5e-97
Alpavirinae_Human_feces_A_016_Microviridae_AG007_putative.pepti... 73.6 4e-19
unnamed protein product 57.8 3e-13
unnamed protein product 56.6 6e-13
unnamed protein product 55.1 3e-12
Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M... 53.5 7e-12
Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.pepti... 49.7 1e-10
unnamed protein product 49.7 1e-10
Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.pepti... 48.5 3e-10
unnamed protein product 48.9 3e-10
> Alpavirinae_Human_gut_33_005_Microviridae_AG0183_putative.peptidase.M15A
Length=131
Score = 273 bits (699), Expect = 5e-97, Method: Compositional matrix adjust.
Identities = 130/131 (99%), Positives = 130/131 (99%), Gaps = 0/131 (0%)
Query 1 MYRPKYFSFTEMILTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRT 60
MYRPKYFSFTEMI TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRT
Sbjct 1 MYRPKYFSFTEMIHTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRT 60
Query 61 PFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYSP 120
PFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYSP
Sbjct 61 PFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYSP 120
Query 121 TYRHEVIVKDN 131
TYRHEVIVKDN
Sbjct 121 TYRHEVIVKDN 131
> Alpavirinae_Human_feces_A_016_Microviridae_AG007_putative.peptidase
Length=109
Score = 73.6 bits (179), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 40/103 (39%), Positives = 57/103 (55%), Gaps = 4/103 (4%)
Query 22 NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAA 81
+N P + I L ++ +ILD VR GKPI+V SG+R LN +VGGV S H +GLAA
Sbjct 5 DNTPTEEVIENLKKVM-YILDIVRAHIGKPILVNSGYRCKRLNEMVGGVQKSMHTKGLAA 63
Query 82 DLRC---DDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYSPT 121
D R +D +F+ + E+ F + K F+H+ S T
Sbjct 64 DFRTGKKEDINTMFEFLKENQKEFKIIELINYKTFIHMGVSET 106
> unnamed protein product
Length=130
Score = 57.8 bits (138), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 42/74 (57%), Gaps = 12/74 (16%)
Query 40 ILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAESD 99
+L+ +R + KP++VTSG+RTP N +GGV SQH +G AAD++ S
Sbjct 43 VLEDIRAHFNKPVIVTSGYRTPEYNTKIGGVKNSQHTKGTAADIKV------------SG 90
Query 100 LPFDQLIYYQKKKF 113
+P ++ Y K K+
Sbjct 91 IPAKEVQKYLKNKY 104
> unnamed protein product
Length=128
Score = 56.6 bits (135), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 21/45 (47%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 40 ILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLR 84
IL+ +RE + +P+++ SG+RTP N+ VGG S H +G+AAD+R
Sbjct 41 ILETIREHFNEPVIINSGYRTPAWNKKVGGAENSYHCKGMAADIR 85
> unnamed protein product
Length=147
Score = 55.1 bits (131), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 57/108 (53%), Gaps = 9/108 (8%)
Query 23 NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAAD 82
N P + L +LC + L+ +R G ++VTSGFR +N VGGV S H+ GLAAD
Sbjct 28 NAPSSYVRDNLYQLCLF-LEQLRSQLGCAVIVTSGFRNVSVNNSVGGVLNSDHLYGLAAD 86
Query 83 LRCDD--PKALFDLIAES---DLPFDQLIYYQKKKFVHVSYSPTYRHE 125
+R P L + I ++ Q+I Y F+HVS + Y+H+
Sbjct 87 IRVKGYTPTKLCNFIRSIPLLNVQVGQVIIY--PTFLHVSIN-RYKHK 131
> Gokush_Human_gut_33_003_Microviridae_AG064_putative.peptidase.M15_3
Length=130
Score = 53.5 bits (127), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 39/64 (61%), Gaps = 2/64 (3%)
Query 35 ELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALF 92
E IL+ R+ KPIV+TSG+RT N+ VGG S H +G+AAD+R + PK L
Sbjct 38 EYLAAILEIARKKINKPIVITSGYRTVSHNKKVGGAKYSYHTRGMAADIRANGITPKELA 97
Query 93 DLIA 96
+++
Sbjct 98 KVLS 101
> Alpavirinae_Human_feces_B_039_Microviridae_AG095_putative.peptidase.M15
Length=102
Score = 49.7 bits (117), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/82 (40%), Positives = 45/82 (55%), Gaps = 8/82 (10%)
Query 54 VTSGFRTPFLNRLVGGVSTSQHMQGLAADLR---CDDPKALFDLI---AESDLPFDQLIY 107
VTS RTP N+ GGV SQH+ G A D++ L ++I +++ PFDQLI
Sbjct 21 VTSARRTPEQNKAAGGVPNSQHLVGEAVDIKPYGSTTFNKLLEMIHFFSDNVSPFDQLII 80
Query 108 YQKKKFVHVSYSPTYRHEVIVK 129
Y F+HVS+ R +VI K
Sbjct 81 Y--PTFIHVSFCSRNRRQVIDK 100
> unnamed protein product
Length=102
Score = 49.7 bits (117), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 45/89 (51%), Gaps = 9/89 (10%)
Query 33 LLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRCDDPK--A 90
L++ WIL R ++ VTS FRT N G TSQH+ G A DL+ D
Sbjct 5 LMKFVEWILH--RNVH---FTVTSAFRTEEQNDACNGSKTSQHLTGDAIDLKPVDSSVDG 59
Query 91 LFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
+I S FDQ+I Y + FVHVS++
Sbjct 60 FLLMIKGSPFKFDQIIKY--RTFVHVSFA 86
> Alpavirinae_Human_gut_22_017_Microviridae_AG0394_putative.peptidase.M15A
Length=102
Score = 48.5 bits (114), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 45/89 (51%), Gaps = 9/89 (10%)
Query 33 LLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRCDDPKA-- 90
L++ W+L R ++ VTS FRT N G SQH+ G A DL+ D
Sbjct 5 LMKFVEWLLR--RNLH---FTVTSAFRTKEQNEACNGSDHSQHLTGDAIDLKPLDFSVDL 59
Query 91 LFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
L +I S FDQLI Y + FVH+S++
Sbjct 60 LVSMIKGSSFKFDQLIKY--RTFVHISFA 86
> unnamed protein product
Length=130
Score = 48.9 bits (115), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 27/85 (32%), Positives = 44/85 (52%), Gaps = 4/85 (5%)
Query 40 ILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADL--RCDDPKALFDLIAE 97
+L+ +RE P+++ SG+RTP N VGG S H++G+AAD+ + K + ++ +
Sbjct 42 VLEIIRESVNAPVIINSGYRTPEWNAKVGGAKCSYHVKGMAADIAVKGHTTKEVAEIASR 101
Query 98 SDLPFDQLIYYQKKKFVHVSYSPTY 122
+I Y FVHV Y
Sbjct 102 ILGNHGGVIRYT--NFVHVDVREGY 124
Lambda K H a alpha
0.323 0.139 0.431 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 8018811