bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_2
Length=351
Score E
Sequences producing significant alignments: (Bits) Value
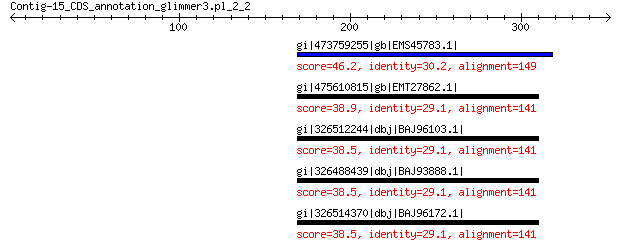
gi|473759255|gb|EMS45783.1| Guanine nucleotide-binding protein s... 46.2 0.037
gi|475610815|gb|EMT27862.1| Puromycin-sensitive aminopeptidase 38.9 6.5
gi|326512244|dbj|BAJ96103.1| predicted protein 38.5 7.4
gi|326488439|dbj|BAJ93888.1| predicted protein 38.5 7.5
gi|326514370|dbj|BAJ96172.1| predicted protein 38.5 8.2
>gi|473759255|gb|EMS45783.1| Guanine nucleotide-binding protein subunit beta [Triticum urartu]
Length=1344
Score = 46.2 bits (108), Expect = 0.037, Method: Composition-based stats.
Identities = 45/164 (27%), Positives = 74/164 (45%), Gaps = 30/164 (18%)
Query 169 KENTEAANYNLALQEQSKVFEEARTMANQNDIIEATKQTQIDTVIQNYYLNSLTAFEKIA 228
KE +EA QE+S++ + +++ ++EA D V + AF +
Sbjct 798 KETSEA-------QEKSRILGSLSSCPDKDIVVEALNLMLTDEV------RNQDAFYVLG 844
Query 229 GIELKGKEAAYISKQIEW---------------YSFEAITKRMSAEAMQKMAESAAERVK 273
GI L+G+EAA+ + W + ++ S E +++E A RVK
Sbjct 845 GISLEGREAAWAWLKDNWDHVVKTWPSSSLISDFVNSTVSPFTSEEKAAEVSEFFATRVK 904
Query 274 NDFEIAGKKIDQEQEKILQTWIFESVKSLCNVAQTTGDIISLFK 317
FE A K+ E+ +I WI +S+KS N+AQT + L K
Sbjct 905 PSFERALKQ-SLERVRISARWI-DSIKSEANLAQTLCSLSRLVK 946
>gi|475610815|gb|EMT27862.1| Puromycin-sensitive aminopeptidase [Aegilops tauschii]
Length=827
Score = 38.9 bits (89), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 71/156 (46%), Gaps = 30/156 (19%)
Query 169 KENTEAANYNLALQEQSKVFEEARTMANQNDIIEATKQTQIDTVIQNYYLNSLTAFEKIA 228
KE +EA QE+S++ + +++ +++A D V + AF +
Sbjct 679 KETSEA-------QEKSRILGSLSSCPDKDIVVQALNLMLTDEV------RNQDAFYVLG 725
Query 229 GIELKGKEAAYISKQIEW---------------YSFEAITKRMSAEAMQKMAESAAERVK 273
GI L+G+EAA+ + W + ++ S E +++E A RVK
Sbjct 726 GISLEGREAAWAWLKDNWDHVVKTWPSSSLISDFVNSTVSPFTSEEKAAEVSEFFATRVK 785
Query 274 NDFEIAGKKIDQEQEKILQTWIFESVKSLCNVAQTT 309
FE A K+ E+ +I WI +S+KS ++AQT
Sbjct 786 PSFERALKQ-SLERVRISARWI-DSIKSEPSLAQTV 819
>gi|326512244|dbj|BAJ96103.1| predicted protein [Hordeum vulgare subsp. vulgare]
gi|326514104|dbj|BAJ92202.1| predicted protein [Hordeum vulgare subsp. vulgare]
gi|326528277|dbj|BAJ93320.1| predicted protein [Hordeum vulgare subsp. vulgare]
Length=871
Score = 38.5 bits (88), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 71/156 (46%), Gaps = 30/156 (19%)
Query 169 KENTEAANYNLALQEQSKVFEEARTMANQNDIIEATKQTQIDTVIQNYYLNSLTAFEKIA 228
KE +EA QE+S++ + +++ ++EA D V + AF +
Sbjct 723 KETSEA-------QEKSRILGSLSSCPDKDIVVEALNLMLTDEV------RNQDAFYVLG 769
Query 229 GIELKGKEAAYISKQIEW---------------YSFEAITKRMSAEAMQKMAESAAERVK 273
GI L+G+EAA+ + W + ++ S E ++++ A RVK
Sbjct 770 GISLEGREAAWAWLKDNWDHVVKTWPSSSLISDFVNSTVSPFTSEEKAAEVSQFFATRVK 829
Query 274 NDFEIAGKKIDQEQEKILQTWIFESVKSLCNVAQTT 309
FE A K+ E+ +I WI +S+KS ++AQT
Sbjct 830 PSFERALKQ-SLERVRISARWI-DSIKSEPSLAQTV 863
>gi|326488439|dbj|BAJ93888.1| predicted protein [Hordeum vulgare subsp. vulgare]
Length=871
Score = 38.5 bits (88), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 71/156 (46%), Gaps = 30/156 (19%)
Query 169 KENTEAANYNLALQEQSKVFEEARTMANQNDIIEATKQTQIDTVIQNYYLNSLTAFEKIA 228
KE +EA QE+S++ + +++ ++EA D V + AF +
Sbjct 723 KETSEA-------QEKSRILGSLSSCPDKDIVVEALNLMLTDEV------RNQDAFYVLG 769
Query 229 GIELKGKEAAYISKQIEW---------------YSFEAITKRMSAEAMQKMAESAAERVK 273
GI L+G+EAA+ + W + ++ S E ++++ A RVK
Sbjct 770 GISLEGREAAWAWLKDNWDHVVKTWPSSSLISDFVNSTVSPFTSEEKAAEVSQFFATRVK 829
Query 274 NDFEIAGKKIDQEQEKILQTWIFESVKSLCNVAQTT 309
FE A K+ E+ +I WI +S+KS ++AQT
Sbjct 830 PSFERALKQ-SLERVRISARWI-DSIKSEPSLAQTV 863
>gi|326514370|dbj|BAJ96172.1| predicted protein [Hordeum vulgare subsp. vulgare]
Length=687
Score = 38.5 bits (88), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 71/156 (46%), Gaps = 30/156 (19%)
Query 169 KENTEAANYNLALQEQSKVFEEARTMANQNDIIEATKQTQIDTVIQNYYLNSLTAFEKIA 228
KE +EA QE+S++ + +++ ++EA D V + AF +
Sbjct 539 KETSEA-------QEKSRILGSLSSCPDKDIVVEALNLMLTDEV------RNQDAFYVLG 585
Query 229 GIELKGKEAAYISKQIEW---------------YSFEAITKRMSAEAMQKMAESAAERVK 273
GI L+G+EAA+ + W + ++ S E ++++ A RVK
Sbjct 586 GISLEGREAAWAWLKDNWDHVVKTWPSSSLISDFVNSTVSPFTSEEKAAEVSQFFATRVK 645
Query 274 NDFEIAGKKIDQEQEKILQTWIFESVKSLCNVAQTT 309
FE A K+ E+ +I WI +S+KS ++AQT
Sbjct 646 PSFERALKQ-SLERVRISARWI-DSIKSEPSLAQTV 679
Lambda K H a alpha
0.310 0.124 0.328 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2072647353324