bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_4
Length=651
Score E
Sequences producing significant alignments: (Bits) Value
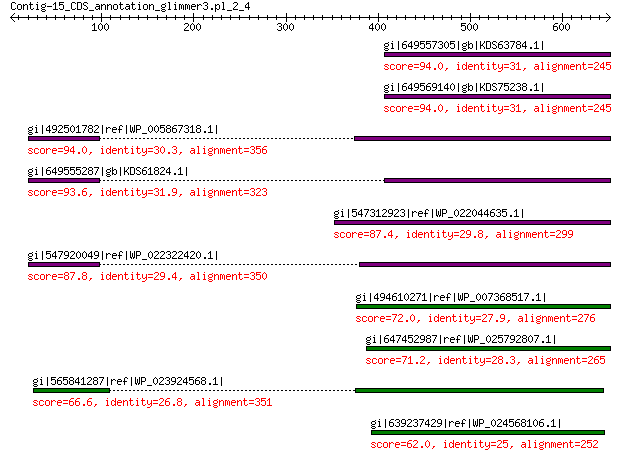
gi|649557305|gb|KDS63784.1| capsid family protein 94.0 6e-18
gi|649569140|gb|KDS75238.1| capsid family protein 94.0 2e-17
gi|492501782|ref|WP_005867318.1| hypothetical protein 94.0 7e-17
gi|649555287|gb|KDS61824.1| capsid family protein 93.6 9e-17
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 87.4 2e-15
gi|547920049|ref|WP_022322420.1| capsid protein VP1 87.8 6e-15
gi|494610271|ref|WP_007368517.1| capsid protein 72.0 6e-10
gi|647452987|ref|WP_025792807.1| hypothetical protein 71.2 1e-09
gi|565841287|ref|WP_023924568.1| hypothetical protein 66.6 3e-08
gi|639237429|ref|WP_024568106.1| hypothetical protein 62.0 9e-07
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 94.0 bits (232), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 76/246 (31%), Positives = 111/246 (45%), Gaps = 19/246 (8%)
Query 407 LNRIAVSGGSYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATEN-EPLGT 465
R A SG Y + + + + R + P F GG I EV+ S+T++ P
Sbjct 18 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 77
Query 466 LAGRGINAGKQKGGKIKIRATEPGYMMCITSITPRIDYSQGNDFDSDWISLDDMHKPALD 525
+AG GI+AG G E GY+M I SI PR Y QG D D + P
Sbjct 78 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFA 135
Query 526 GIGYQDSVNSGRAWWDDVYINNTGKAAKRTAGKTVAWIDYMTNVNRTYGNFAAGMSEAFM 585
+G Q+ N +++Y+N + A + T G T + +Y + N +G+F M AF
Sbjct 136 HLGEQEIKN------EELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNM--AFW 187
Query 586 VLNRNYEMKYEAGTNPKISDLTTYIDPVKYNYIFADTSIDAMNFWVQIKFDITARRLMSA 645
LNR ++ K P ++ TT+++ N +FA +WVQI DI A RLM
Sbjct 188 HLNRIFKEK------PNLN--TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPK 239
Query 646 KQIPNL 651
P L
Sbjct 240 YGTPML 245
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 94.0 bits (232), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 76/246 (31%), Positives = 111/246 (45%), Gaps = 19/246 (8%)
Query 407 LNRIAVSGGSYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATEN-EPLGT 465
R A SG Y + + + + R + P F GG I EV+ S+T++ P
Sbjct 163 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 222
Query 466 LAGRGINAGKQKGGKIKIRATEPGYMMCITSITPRIDYSQGNDFDSDWISLDDMHKPALD 525
+AG GI+AG G E GY+M I SI PR Y QG D D + P
Sbjct 223 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFA 280
Query 526 GIGYQDSVNSGRAWWDDVYINNTGKAAKRTAGKTVAWIDYMTNVNRTYGNFAAGMSEAFM 585
+G Q+ N +++Y+N + A + T G T + +Y + N +G+F M AF
Sbjct 281 HLGEQEIKN------EELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNM--AFW 332
Query 586 VLNRNYEMKYEAGTNPKISDLTTYIDPVKYNYIFADTSIDAMNFWVQIKFDITARRLMSA 645
LNR ++ K P ++ TT+++ N +FA +WVQI DI A RLM
Sbjct 333 HLNRIFKEK------PNLN--TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPK 384
Query 646 KQIPNL 651
P L
Sbjct 385 YGTPML 390
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 94.0 bits (232), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 80/279 (29%), Positives = 124/279 (44%), Gaps = 19/279 (7%)
Query 374 DGANGINEITAVDVSDGTLSMDALNLQQKVYNMLNRIAVSGGSYRDWLETVYASGQYIER 433
DGA + V+V + +S++ L + R A SG Y + + + + R
Sbjct 278 DGAYLEPDNFQVNVDELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDAR 337
Query 434 CETPTFEGGTSQEIVFQEVVSNSATEN-EPLGTLAGRGINAGKQKGGKIKIRATEPGYMM 492
+ P F GG I EV+ SAT++ P +AG GI+AG G K E GY++
Sbjct 338 LQRPQFLGGGRTPISVSEVLQTSATDSTSPQANMAGHGISAGVNHG--FKRYFEEHGYII 395
Query 493 CITSITPRIDYSQGNDFDSDWISLDDMHKPALDGIGYQDSVNSGRAWWDDVYINNTGKAA 552
I SI PR Y QG D D + P +G Q+ N ++VY+ T +
Sbjct 396 GIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKN------EEVYLQQTPASN 449
Query 553 KRTAGKTVAWIDYMTNVNRTYGNFAAGMSEAFMVLNRNYEMKYEAGTNPKISDLTTYIDP 612
T G T + +Y ++N +G+F M AF LNR + +P ++ TT+++
Sbjct 450 NGTFGYTPRYAEYKYSMNEVHGDFRGNM--AFWHLNRIFS------ESPNLN--TTFVEC 499
Query 613 VKYNYIFADTSIDAMNFWVQIKFDITARRLMSAKQIPNL 651
N +FA +W+Q+ D+ A RLM P L
Sbjct 500 NPSNRVFATAETSDDKYWIQLYQDVKALRLMPKYGTPML 538
Score = 47.4 bits (111), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 37/79 (47%), Gaps = 1/79 (1%)
Query 20 VDLKTYNRSTHNLSTVIRNTQSAGTLVPTCTIVMQKDDTFEINIESSVLTHPTTGPLYGS 79
V LK R+ NLS + T +AG LVP + D F +N E V P P+
Sbjct 8 VKLKRPRRNVFNLSYENKLTANAGELVPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHR 67
Query 80 FKLENHVFFIPFR-LYNSW 97
+ H FF+P R L+N W
Sbjct 68 VDVFTHYFFVPNRLLWNQW 86
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 93.6 bits (231), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 76/246 (31%), Positives = 111/246 (45%), Gaps = 19/246 (8%)
Query 407 LNRIAVSGGSYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATEN-EPLGT 465
R A SG Y + + + + R + P F GG I EV+ S+T++ P
Sbjct 314 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 373
Query 466 LAGRGINAGKQKGGKIKIRATEPGYMMCITSITPRIDYSQGNDFDSDWISLDDMHKPALD 525
+AG GI+AG G E GY+M I SI PR Y QG D D + P
Sbjct 374 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFA 431
Query 526 GIGYQDSVNSGRAWWDDVYINNTGKAAKRTAGKTVAWIDYMTNVNRTYGNFAAGMSEAFM 585
+G Q+ N +++Y+N + A + T G T + +Y + N +G+F M AF
Sbjct 432 HLGEQEIKN------EELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNM--AFW 483
Query 586 VLNRNYEMKYEAGTNPKISDLTTYIDPVKYNYIFADTSIDAMNFWVQIKFDITARRLMSA 645
LNR ++ K P ++ TT+++ N +FA +WVQI DI A RLM
Sbjct 484 HLNRIFKEK------PNLN--TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPK 535
Query 646 KQIPNL 651
P L
Sbjct 536 YGTPML 541
Score = 45.8 bits (107), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 37/79 (47%), Gaps = 1/79 (1%)
Query 20 VDLKTYNRSTHNLSTVIRNTQSAGTLVPTCTIVMQKDDTFEINIESSVLTHPTTGPLYGS 79
V LK R+ NLS + T +AG L+P + D F +N E V P P+
Sbjct 8 VKLKRPRRNVFNLSYENKLTVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHR 67
Query 80 FKLENHVFFIPFRL-YNSW 97
+ H FF+P RL +N W
Sbjct 68 VDVFTHYFFVPNRLIWNKW 86
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 87.4 bits (215), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 89/333 (27%), Positives = 138/333 (41%), Gaps = 41/333 (12%)
Query 353 GLLLKTYNSDLYQNWINTEWIDGANGINEITAVDVSDGT---LSMDALNLQQKVYNMLNR 409
GLL Y+ DL+ N I + A I + A+D++ T +++ L L+ K+ N ++R
Sbjct 11 GLLSVPYSPDLFGNIIK-QGSSPAVEIEVMNALDLNISTGFSVAVPELRLRTKIQNWMDR 69
Query 410 IAVSGGSYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATENEPLGTLAGR 469
+ VSGG D T++ + P F G V+Q ++ S G+ +G
Sbjct 70 LFVSGGRVGDVFRTLWGTKSSAIYVNKPDFLG------VWQASINPSNVRAMANGSASGE 123
Query 470 GINAGKQKG-----------GKIKIRATEPGYMMCITSITPRIDYSQGNDFDSDWISLDD 518
N G+ I A EPG M IT + P YSQG D IS D
Sbjct 124 DANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFGD 183
Query 519 MHKPALDGIGYQDSVN------------SGRAWWDDVYINNTGKAA-----KRTAGKTVA 561
P L+GIG+Q +G + +TG + G+ VA
Sbjct 184 DFNPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEVA 243
Query 562 WIDYMTNVNRTYGNFAAGMSEAFMVLNRNYEMKYEAGTNPKISD---LTTYIDPVKYNYI 618
W T+ +R +G+FA + + VL R + + D TYI+P+ + Y+
Sbjct 244 WSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYTGTYINPLDWQYV 303
Query 619 FADTSIDAMNFWVQIKFDITARRLMSAKQIPNL 651
F D ++ A NF FD+ +SA +P L
Sbjct 304 FVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 87.8 bits (216), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 78/273 (29%), Positives = 118/273 (43%), Gaps = 19/273 (7%)
Query 380 NEITAVDVSDGTLSMDALNLQQKVYNMLNRIAVSGGSYRDWLETVYASGQYIERCETPTF 439
N V+V + ++++ L + R A G Y + + + + R + P F
Sbjct 299 NGTLKVNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQF 358
Query 440 EGGTSQEIVFQEVVSNSAT-ENEPLGTLAGRGINAGKQKGGKIKIRATEPGYMMCITSIT 498
GG I EV+ S+T E P +AG GI+AG G K E GY++ I SIT
Sbjct 359 LGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGINNG--FKHYFEEHGYIIGIMSIT 416
Query 499 PRIDYSQGNDFDSDWISLDDMHKPALDGIGYQDSVNSGRAWWDDVYINNTGKAAKRTAGK 558
PR Y QG D D + P + Q+ N +D NN T G
Sbjct 417 PRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNG------TFGY 470
Query 559 TVAWIDYMTNVNRTYGNFAAGMSEAFMVLNRNYEMKYEAGTNPKISDLTTYIDPVKYNYI 618
T + +Y + + +G+F +S F LNR +E K P ++ TT+++ N +
Sbjct 471 TPRYAEYKYHPSEAHGDFRGNLS--FWHLNRIFEDK------PNLN--TTFVECKPSNRV 520
Query 619 FADTSIDAMNFWVQIKFDITARRLMSAKQIPNL 651
FA + + FWVQ+ D+ A RLM P L
Sbjct 521 FATSETEDDKFWVQMYQDVKALRLMPKYGTPML 553
Score = 45.4 bits (106), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/79 (32%), Positives = 37/79 (47%), Gaps = 1/79 (1%)
Query 20 VDLKTYNRSTHNLSTVIRNTQSAGTLVPTCTIVMQKDDTFEINIESSVLTHPTTGPLYGS 79
+ +K R+ NLS + T + G LVP + + D F + ES V P P+
Sbjct 8 IRMKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMHR 67
Query 80 FKLENHVFFIPFRL-YNSW 97
+ H FF+P RL +N W
Sbjct 68 VNVFTHYFFVPNRLVWNEW 86
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 72.0 bits (175), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 77/305 (25%), Positives = 126/305 (41%), Gaps = 32/305 (10%)
Query 376 ANGINEITAVDVSDGTLSMDALNLQQKVYNMLNRIAVSGG-SYRDWLETVYASGQYIERC 434
+NG++ + LS++ L + ML + G Y +E + R
Sbjct 230 SNGVSGASTFINGVSVLSVNDLRAAFALDKMLEATRRANGLDYSSQIEAHFGFRVPESRA 289
Query 435 ETPTFEGGTSQEIVFQEVVSNS-----ATENEPLGTLAGRGINAGKQKGGKIKIRATEPG 489
F GG +V EVV+ S A E+ LG L G+G+ G I E G
Sbjct 290 GDARFIGGFDNPVVISEVVNQSEFDRGADESPCLGDLGGKGV--GSLNSSSIDFDVKEHG 347
Query 490 YMMCITSITPRIDYSQGNDFD--SDWISLDDMHKPALDGIGYQDSVNSG--RAWWDDVYI 545
+MCI S+ P+ +Y G FD + + +D +P +GYQ V S + D+
Sbjct 348 IIMCIYSVVPQTEY-NGTYFDPFNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDNPVP 406
Query 546 NNTGKAAKRTAGKTVAWID--------------YMTNVNRTYGNFAAGMSEAFMVLNR-N 590
+ K + AG ++ I+ Y T+ + +G F +G+S ++ R +
Sbjct 407 DGPEKQKRLAAGYPLSSIEANNRLLGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSPRYD 466
Query 591 YEMKYEAG----TNPKISDLTTYIDPVKYNYIFADTSIDAMNFWVQIKFDITARRLMSAK 646
+ +AG N S Y++P N IF +++ A +F V FD+ A R MS
Sbjct 467 FGFDGKAGDKKLVNSPWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDVKAVRPMSVS 526
Query 647 QIPNL 651
+ L
Sbjct 527 GLAGL 531
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 71.2 bits (173), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 75/302 (25%), Positives = 127/302 (42%), Gaps = 47/302 (16%)
Query 387 VSDGTLSMDALNLQQKVYNMLNRIAVSGG-SYRDWLETVYASGQYIERCETPTFEGGTSQ 445
VS + S++ L + ML + G Y +E + R F GG
Sbjct 293 VSPSSFSVNDLRAAFALDKMLEATRRANGLDYASQIEAHFGFKVPESRANDARFLGGFDN 352
Query 446 EIVFQEVVS---NSATE--NEPLGTLAGRGINAGKQKGGKIKIRATEPGYMMCITSITPR 500
IV EVVS N+A++ + +G L G+GI G G I+ +TE G +MCI S+ P+
Sbjct 353 SIVVSEVVSTNGNAASDGSHASIGDLGGKGI--GSMSSGTIEFDSTEHGIIMCIYSVAPQ 410
Query 501 IDYSQG--NDFDSDWISLDDMHKPALDGIGYQD-----------SVNSGRAWWDDVYINN 547
+Y+ + F+ ++ + ++P +GYQ +N +A + D+ +NN
Sbjct 411 SEYNASYLDPFNRK-LTREQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDIELNN 469
Query 548 TGKAAKRTAGKTVAWIDYMTNVNRTYGNFAAGMSEAFMVLNRNYEMKY------------ 595
G V + +Y T + +G+F +G S ++ R ++ Y
Sbjct 470 N------LLGYQVRYNEYKTARDLVFGDFESGKSLSYWCTPR-FDFGYGDTEKKIAPENK 522
Query 596 ------EAGTNPKISDLTTYIDPVKYNYIFADTSIDAMNFWVQIKFDITARRLMSAKQIP 649
+ G S YI+P N IF +++ A +F V D+ A R MS +
Sbjct 523 GGADYRKKGNRSHWSSRNFYINPNLVNPIFLTSAVQADHFIVNSFLDVKAVRPMSVTGLS 582
Query 650 NL 651
+L
Sbjct 583 SL 584
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 66.6 bits (161), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 72/283 (25%), Positives = 120/283 (42%), Gaps = 32/283 (11%)
Query 375 GANGINEITAVDVSDGTLSMDALNLQQKVYNMLNRI-AVSGGSYRDWLETVYASGQYIER 433
G+N I+ I+ D+ +M AL ML R A +G Y + + + R
Sbjct 384 GSNNISYISPNDIR----AMFALE------KMLERTRAANGLDYSNQIAAHFGFKVPESR 433
Query 434 CETPTFEGGTSQEIVFQEVVSNS-------ATENEPLGTLAGRGINAGKQKGGKIKIRAT 486
+F GG +I EVV+ S A+ +G + G+GI G G I
Sbjct 434 KNCASFIGGFDNQISISEVVTTSNGSVDGTASTGSVVGQVFGKGI--GAMNSGHISYDVK 491
Query 487 EPGYMMCITSITPRIDYS--QGNDFDSDWISLDDMHKPALDGIGYQDSVNSGRAWWDDVY 544
E G +MCI SI P++DY + + F+ + S +D +P + +G Q + S +
Sbjct 492 EHGLIMCIYSIAPQVDYDARELDPFNRKF-SREDYFQPEFENLGMQPVIQSDLCLCINSA 550
Query 545 INNTGKAAKRTAGKTVAWIDYMTNVNRTYGNFAAGMS-EAFMVLNRNYEMKYEAGTNPKI 603
+++ G + +++Y T + +G F +G S A+ NY ++ + P +
Sbjct 551 KSDSSDQHNNVLGYSARYLEYKTARDIIFGEFMSGGSLSAWATPKNNYTFEFGKLSLPDL 610
Query 604 SDLTTYIDPVKYNYIFA---DTSIDAMNFWVQIKFDITARRLM 643
+DP IFA + S+ F V FD+ A R M
Sbjct 611 -----LVDPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIRPM 648
Score = 42.7 bits (99), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 22/82 (27%), Positives = 40/82 (49%), Gaps = 2/82 (2%)
Query 26 NRSTHNLSTVIRNTQSAGTLVPTCTIVMQKDDTFEINIESSVLTHPTTGPLYGSFKLENH 85
NR+ ++LS+ + AG L+P T + F I+++ V P + K H
Sbjct 14 NRNGYDLSSRRIFSAPAGALLPIATWEANPGEKFRISVQDLVRAQPLNTAAFARCKEYYH 73
Query 86 VFFIPFRLYNSWLHNNRLGIGL 107
FF+P++ + W H++R G+
Sbjct 74 FFFVPYK--SLWQHSDRFFTGV 93
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 62.0 bits (149), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 63/254 (25%), Positives = 112/254 (44%), Gaps = 17/254 (7%)
Query 393 SMDALNLQQKVYNMLNRIAVSGGSYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEV 452
+++ L K+ L + A +G Y + + + + R + P F GG I+ EV
Sbjct 299 TINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILISEV 358
Query 453 VSNSATEN-EPLGTLAGRGINAGKQKGGKIKIRATEPGYMMCITSITPRIDYSQGNDFDS 511
+ S+T++ P G +AG GI+ GK+ GG K E GY++ + S+ P+ YSQG
Sbjct 359 LQQSSTDSTTPQGNMAGHGISVGKE-GGFSKF-FEEHGYVIGLMSVIPKTSYSQGIPRHF 416
Query 512 DWISLDDMHKPALDGIGYQDSVNSGRAWWDDVYINNTGKA-AKRTAGKTVAWIDYMTNVN 570
D P + IG Q N +++ N G + G + +Y + +
Sbjct 417 SKFDKFDYFWPQFEHIGEQPVYNK------EIFAKNVGDYDSGGVFGYVPRYSEYKYSPS 470
Query 571 RTYGNFAAGMSEAFMVLNRNYEMKYEAGTNPKISDLTTYIDPVKYNYIFADTSIDAMNFW 630
+G+F + F L R +++ PK++ ++ + IFA ++ F+
Sbjct 471 TIHGDFKDTL--YFWHLGR----IFDSSAPPKLNRDFIEVNKSGLSRIFA-VEDNSDKFY 523
Query 631 VQIKFDITARRLMS 644
+ ITA+R MS
Sbjct 524 CHLYQKITAKRKMS 537
Lambda K H a alpha
0.315 0.133 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4943415339264