bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_5
Length=82
Score E
Sequences producing significant alignments: (Bits) Value
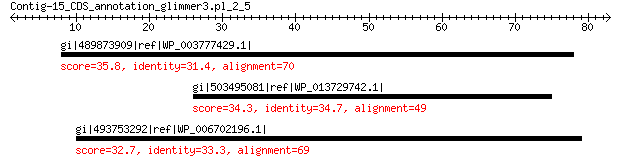
gi|489873909|ref|WP_003777429.1| hypothetical protein 35.8 0.49
gi|503495081|ref|WP_013729742.1| DNA helicase 34.3 7.0
gi|493753292|ref|WP_006702196.1| glycine cleavage system protein H 32.7 7.8
>gi|489873909|ref|WP_003777429.1| hypothetical protein [Alloiococcus otitis]
gi|425730934|gb|EKU93764.1| hypothetical protein HMPREF9698_00712 [Alloiococcus otitis ATCC
51267]
Length=110
Score = 35.8 bits (81), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 22/72 (31%), Positives = 41/72 (57%), Gaps = 2/72 (3%)
Query 8 FKIRLLPESEEYVITIG--SHLATEEKFKSEKAAQMQINKTNWDLVSALVYALKEADEWA 65
+ I + PE ++ V T+G + ++ + + A ++ +KT D++S L + E A
Sbjct 17 YVISMTPEMQDDVGTVGFVEFVDKDDIKEGDTLANIEASKTVLDVLSPLAGRIVARHEEA 76
Query 66 EKNPELINQTKE 77
EKNPEL+N +K+
Sbjct 77 EKNPELLNSSKQ 88
>gi|503495081|ref|WP_013729742.1| DNA helicase [Mycoplasma mycoides]
gi|331703646|ref|YP_004400333.1| Replicative DNA helicase DnaC [Mycoplasma mycoides subsp. capri
LC str. 95010]
gi|328802201|emb|CBW54355.1| Replicative DNA helicase DnaC [Mycoplasma mycoides subsp. capri
LC str. 95010]
Length=437
Score = 34.3 bits (77), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 17/49 (35%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 26 HLATEEKFKSEKAAQMQINKTNWDLVSALVYALKEADEWAEKNPELINQ 74
+LA FK K A + +N L++ L+ + + D A KNP++INQ
Sbjct 215 NLALNAAFKEHKVALFSLEMSNEQLIARLLSRISKVDSLAFKNPKIINQ 263
>gi|493753292|ref|WP_006702196.1| glycine cleavage system protein H [Facklamia ignava]
gi|405579715|gb|EKB53810.1| hypothetical protein HMPREF9707_01563 [Facklamia ignava CCUG
37419]
Length=110
Score = 32.7 bits (73), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 10 IRLLPESEEYVITIGSHLATEEK-FKSEKA-AQMQINKTNWDLVSALVYALKEADEWAEK 67
I + PE ++ + T+G TEEK K E A A ++ +KT +++S + + E +E A
Sbjct 19 ISMTPELQDDIGTVGFVEFTEEKQLKKEDAIAHIEASKTVLEIISPVTGTVVEINEAAID 78
Query 68 NPELINQTKEA 78
NP+++N A
Sbjct 79 NPKMLNSADTA 89
Lambda K H a alpha
0.307 0.123 0.334 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434005500390