bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-17_CDS_annotation_glimmer3.pl_2_5
Length=511
Score E
Sequences producing significant alignments: (Bits) Value
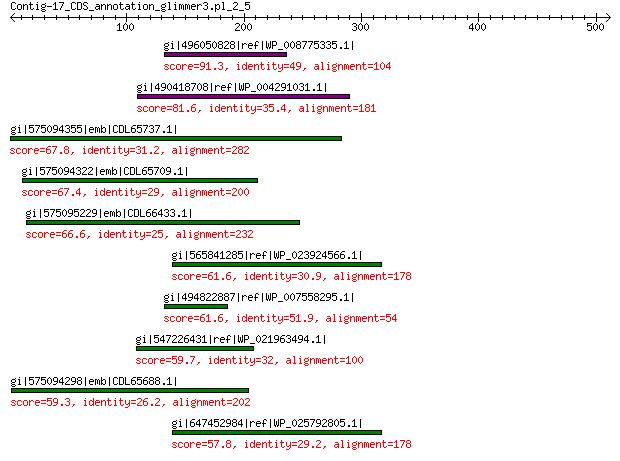
gi|496050828|ref|WP_008775335.1| hypothetical protein 91.3 3e-16
gi|490418708|ref|WP_004291031.1| hypothetical protein 81.6 2e-13
gi|575094355|emb|CDL65737.1| unnamed protein product 67.8 9e-09
gi|575094322|emb|CDL65709.1| unnamed protein product 67.4 1e-08
gi|575095229|emb|CDL66433.1| unnamed protein product 66.6 2e-08
gi|565841285|ref|WP_023924566.1| hypothetical protein 61.6 6e-07
gi|494822887|ref|WP_007558295.1| hypothetical protein 61.6 7e-07
gi|547226431|ref|WP_021963494.1| predicted protein 59.7 3e-06
gi|575094298|emb|CDL65688.1| unnamed protein product 59.3 4e-06
gi|647452984|ref|WP_025792805.1| hypothetical protein 57.8 1e-05
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 91.3 bits (225), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 73/104 (70%), Gaps = 1/104 (1%)
Query 132 IPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHFHLLLFTNSDEVAE 191
+P+L D+Q ++KRLR Y+ K S E + ++AVGEYGPVHFRPH+HLLLF SDE +
Sbjct 117 VPYLRKTDLQLFLKRLRYYVTKQKPS-EKVRYFAVGEYGPVHFRPHYHLLLFLQSDEALQ 175
Query 192 VLRQCHDKSWKFGRSDFQRsaggsasyvssyvNSLCSAPLLYRS 235
+ + K+W FGR D Q S G ++YV+SYVNS C+ P ++++
Sbjct 176 ICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKA 219
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 81.6 bits (200), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 64/190 (34%), Positives = 95/190 (50%), Gaps = 15/190 (8%)
Query 109 IHKTQALGKTDYPVAE------QYGRDNLIPFLNYVDVQNYIKRLRKYLFKVLGSYESLH 162
+ + LG+ D + E ++ +P+L D+Q + KR R Y+ K E +
Sbjct 12 VETGEYLGEADLSIKEIERLQEKFHLFGYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVR 70
Query 163 FYAVGEYGPVHFRPHFHLLLFTNSDEVAEVLRQCHDKSWKFGRSDFQRsaggsasyvssy 222
++A+GEYGPVHFRPH+H+LLF SDE +V + ++W FGR D Q S G +SYV+ Y
Sbjct 71 YFAIGEYGPVHFRPHYHILLFLQSDEALQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGY 130
Query 223 vNSLCSAP---LLYRSCRAFRPKSRASVGFFEKGCDFVEDEDPYAQIEKKIDSVVNGRCY 279
VNS P L C + GF + V P +++ I V+NGR
Sbjct 131 VNSSVLVPKVLTLPTLCPFCVHSQKLGQGFLQSERAKVYSLTPEQFVKRSI--VINGRYK 188
Query 280 NFNGVSVWST 289
F+ VW +
Sbjct 189 EFD---VWRS 195
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 67.8 bits (164), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 88/313 (28%), Positives = 129/313 (41%), Gaps = 89/313 (28%)
Query 1 MRVKTAGSAFKYSYFVTLTYDNEHIPLMRCKVLHSEYEDVVGISGDIHFGDEYHHYIPVS 60
++++ S K+ F TLTY N +IP + +P +
Sbjct 46 LQIQLEASQHKFCIFGTLTYANTYIPRLSL--------------------------VPYN 79
Query 61 EYQCDDSSALRHIFFEQVQGTVPYDREIKEYVPVKDNWFLSIDAIRSFIHKTQALGKTDY 120
+ F V G D+E EY+ D+ S D + S + K G
Sbjct 80 DKT-----------FGVVNGYEMCDKETGEYLGYLDS--PSYD-VESLLDKLHLFGD--- 122
Query 121 PVAEQYGRDNLIPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHFHL 180
+P+L D+Q +IKRLRK L K S + ++A+GEYGPVHFRPH+H
Sbjct 123 -----------VPYLRKRDLQLFIKRLRKNLSKY--SDAKVRYFAMGEYGPVHFRPHYHF 169
Query 181 LLF----------------------------TNSDEVAEVLRQCHDKSWKFGRSDFQRsa 212
LLF + ++ V+ C SWKFGR D Q S
Sbjct 170 LLFFDEIKFTAPSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWKFGRVDAQYSK 229
Query 213 ggsasyvssyvNSLCSAPLLYR--SCRAFRPKSR-ASVGFFEKGCDFVEDEDPYAQIEKK 269
G +A YVSSYV+ S P +Y+ S R F SR GF C+ V + +++
Sbjct 230 GDAAQYVSSYVSGSGSLPKVYQVSSARPFSLHSRFLGQGFLAHECEKVYETPVRDFVKRS 289
Query 270 IDSVVNGRCYNFN 282
++ +NG +FN
Sbjct 290 VE--LNGSNKDFN 300
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 67.4 bits (163), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 58/215 (27%), Positives = 92/215 (43%), Gaps = 51/215 (24%)
Query 11 KYSYFVTLTYDNEHIPLMRCKVLHSEYEDVVGISGDIHFGDEYHHYIPVSEYQCDDSSAL 70
KY YF+TLTYD++++PL VG+ E+ P SE +DS
Sbjct 52 KYCYFLTLTYDDDNLPLFS-----------VGLDT---CATEFVRIYPYSERLRNDS--- 94
Query 71 RHIFFEQVQGTVPYDREIKEYVPVKDNWFLSIDAIRSFIHKTQALGKTDYPVAEQYGRDN 130
+ +D + + + ++ ++ + S HK+ G
Sbjct 95 --FISDFCSDLHNFDNDFVDKMDYYSDYVINYE---SKYHKSCVYGH------------G 137
Query 131 LIPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHFHLLLFTNSDEVA 190
L L Y D+Q ++KRLRK+++K G E + FY +GEYG RPH+H LLF NS ++
Sbjct 138 LYALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEYGTKSLRPHWHCLLFFNSSSLS 195
Query 191 EVLRQCHDKS---------------WKFGRSDFQR 210
+ C + W+FG D +R
Sbjct 196 QAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKR 230
>gi|575095229|emb|CDL66433.1| unnamed protein product [uncultured bacterium]
Length=510
Score = 66.6 bits (161), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 58/234 (25%), Positives = 108/234 (46%), Gaps = 28/234 (12%)
Query 15 FVTLTYDNEHIPLMRCKVLHSEYEDVVGISGDIHFGDEYHHYIPVSEYQCDDSSALRHIF 74
F+ LTYD EH+PL+R IS H D ++ P+++ + + + F
Sbjct 58 FIMLTYDKEHLPLVR-------------ISK--HDFDAMYYKKPINKPEYEKRN-----F 97
Query 75 FEQVQGTVPYDREIKEYVPVKDNWFLSIDAIRSFIHKTQALGKTDYPVAEQYGRDNLIPF 134
F Q+ Y++++ + + + + L ++ Y + ++P
Sbjct 98 FCQLS----YEKQLSKITSLSNRKVFKSAYSSQSGYSMSTLFESGYNNSVHTDCYYMLPT 153
Query 135 LNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHFHLLLFTNSDEVAEVLR 194
L YVDV ++KRLR + + +G ++ F A GEYGP FRPH+H+++ S+ + +
Sbjct 154 LRYVDVSGFLKRLRTRVQREIGE-SNIRFAACGEYGPRGFRPHYHIIVICQSEAARQSVM 212
Query 195 QCHDKSWKFGRSDFQRsaggsasyvssyvNSLCSAPLLYR--SCRAFRPKSRAS 246
+ + W +G S + S + N + +PLL + + + FRP R+S
Sbjct 213 RNYRTCWLYGLSS-AKLYIKSKNSADYVSNYVTCSPLLPKLYTYKPFRPFFRSS 265
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 61.6 bits (148), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 55/192 (29%), Positives = 88/192 (46%), Gaps = 27/192 (14%)
Query 139 DVQNYIKRLRK---YLFKV--LGSYESLHFYAVGEYGPVHFRPHFHLLLFTNSDEVAEVL 193
D+ + KRLR Y FK + + E + ++ EYGP RPH+H +++ +S+EVA V+
Sbjct 119 DIVKFFKRLRSKLSYYFKKHHIITNEKIRYFVCSEYGPKTLRPHYHAIIWFDSEEVARVI 178
Query 194 RQCHDKSWKFGRSDFQ--RsaggsasyvssyvNSLCSAPLLYRSCRAFRPKSRA-SVGFF 250
+ SW G +DF+ S NS+ L + +CR F +S+A SVG+
Sbjct 179 EKMLSSSWSNGFTDFEYVNSTAPQYVAKYVSGNSVLPEILQHDACRTFHLQSQAPSVGY- 237
Query 251 EKGCDFVEDEDPYAQIEKKIDSVVNGRCYN------FNGVSVWSTPPMSYVRTLLPRFSS 304
D Y + EK+ V++G CY + SV+ PP + P+
Sbjct 238 --------RSDDYEKFEKE---VIDG-CYGHFEYDSSSQSSVFVQPPGTLETRCFPKCRE 285
Query 305 ARNDDSTAIIRI 316
R+ +RI
Sbjct 286 YRSLSRIEKLRI 297
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 61.6 bits (148), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/54 (52%), Positives = 40/54 (74%), Gaps = 2/54 (4%)
Query 132 IPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHFHLLLFTN 185
P+L+ ++Q ++KRLRKYL K G + + F+A GEYGP+ FRPHFH+LLF +
Sbjct 118 FPYLSKRELQLFMKRLRKYLDKYEG--QKIRFFATGEYGPLSFRPHFHILLFVD 169
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 59.7 bits (143), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 54/100 (54%), Gaps = 2/100 (2%)
Query 108 FIHKTQALGKTDYPVAEQYGRDNLIPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVG 167
+ HK +L + + D + + + D Q ++KR+RK L K S E + +Y V
Sbjct 106 YWHKCPSLDTYVLMLTAKCNLDGYLSYTSKRDAQLFLKRVRKNLSKY--SDEKIRYYIVS 163
Query 168 EYGPVHFRPHFHLLLFTNSDEVAEVLRQCHDKSWKFGRSD 207
EYGP FR H+H+L F + + +V+ + ++W+FGR D
Sbjct 164 EYGPKTFRAHYHVLFFYDEVKTQKVMSKVIRQAWQFGRVD 203
>gi|575094298|emb|CDL65688.1| unnamed protein product [uncultured bacterium]
Length=478
Score = 59.3 bits (142), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 53/206 (26%), Positives = 88/206 (43%), Gaps = 29/206 (14%)
Query 2 RVKTAGSAFKYSYFVTLTYDNEHIPLMRCKVLHSEYEDVVGISGDIHFGDEYHHY-IPVS 60
+++ S+ +FVTL YDN HIP++ H Y S HF +E +PV
Sbjct 38 KIRNNQSSELSCFFVTLNYDNNHIPVI---FKHDVYN--YNSSDVYHFDEERKELCLPVD 92
Query 61 EYQCDDSSALRHIFFEQVQGTVP-YDREIKEY-VPVKDNWFLSIDAIRSFIHKTQALGKT 118
Y +G P + +I + P+ LS D + S + + KT
Sbjct 93 LY----------------RGVCPAFSNKIDTFNFPLNR---LSTDVVSSLDNHCGVVVKT 133
Query 119 DYPVAEQYGRDNLIPFLNYVDVQNYIKRLRKYLFKVLGSYESLHFYAVGEYGPVHFRPHF 178
+ + + D+Q + KRLR+ L++ G + ++ EYGP +R HF
Sbjct 134 KNHKPVLFNEE-IFSVCYTKDIQLFFKRLRQSLYRKFGFRPFIQYFQTSEYGPTTYRAHF 192
Query 179 HLLLFTNSDEVA-EVLRQCHDKSWKF 203
HL +F E++ + R+ K+W F
Sbjct 193 HLCIFVKRSEISFDSFRKACVKAWPF 218
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 57.8 bits (138), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 52/184 (28%), Positives = 81/184 (44%), Gaps = 13/184 (7%)
Query 139 DVQNYIKRLRK---YLFKVLGSYESLHFYAVGEYGPVHFRPHFHLLLFTNSDEVAEVLRQ 195
DVQ++ KRLR Y K G+ + ++ EYGP FRPH+H +L+ +S+ + L
Sbjct 126 DVQDFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEILHNELNV 185
Query 196 CHDKSWKFGRSDFQRsaggsasyvssyvNSLCSAPLLYRS--CRAFRPKSRASVGFFEKG 253
++WK G +DF ++ YV+ YVN C P R+ F S+ + K
Sbjct 186 LIRETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFLRTEFTSTFHLASKHPCIGYGK- 244
Query 254 CDFVEDEDPYAQIEKKIDSVVNGRCYNFNGVSV-WSTPPMSYVRTLLPRFSSARNDDSTA 312
+D A E I+ C N + + PP S +LP+ R +
Sbjct 245 ------DDEEALYENVINGTYGRNCLNKSTNEFEFVCPPRSLENRILPKCKGYRRISHSE 298
Query 313 IIRI 316
+RI
Sbjct 299 RVRI 302
Lambda K H a alpha
0.324 0.139 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3603121648380