bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_8
Length=222
Score E
Sequences producing significant alignments: (Bits) Value
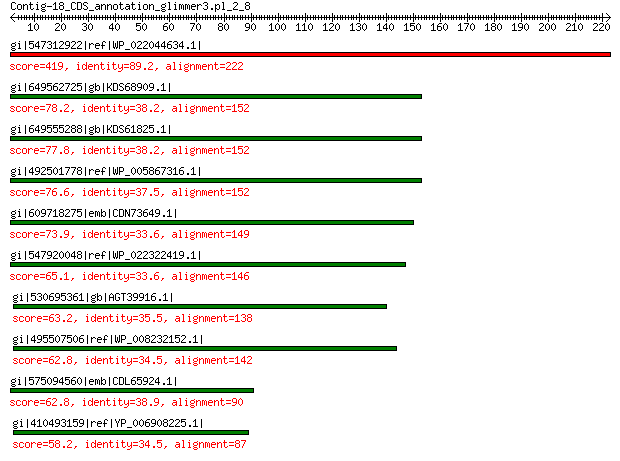
gi|547312922|ref|WP_022044634.1| putative replication initiation... 419 7e-145
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 78.2 3e-14
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 77.8 6e-14
gi|492501778|ref|WP_005867316.1| hypothetical protein 76.6 1e-13
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 73.9 1e-12
gi|547920048|ref|WP_022322419.1| putative replication protein 65.1 2e-09
gi|530695361|gb|AGT39916.1| replication initiator 63.2 1e-08
gi|495507506|ref|WP_008232152.1| hypothetical protein 62.8 1e-08
gi|575094560|emb|CDL65924.1| unnamed protein product 62.8 2e-08
gi|410493159|ref|YP_006908225.1| replication-associated protein 58.2 5e-07
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 419 bits (1077), Expect = 7e-145, Method: Compositional matrix adjust.
Identities = 198/222 (89%), Positives = 208/222 (94%), Gaps = 0/222 (0%)
Query 1 LFLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASR 60
LFLDR RK Y KQIRHWFVCEFGTL+GRPHYHGILF+VPQ LIDGY D+PGHHPLLAS
Sbjct 99 LFLDRFRKVYGKQIRHWFVCEFGTLHGRPHYHGILFNVPQALIDGYDSDMPGHHPLLASC 158
Query 61 WKYGFVFVGYVSEETCSYITKYVTKSINGDKVRPRIISSFGIGSNYLDTEEAVLHKLGNQ 120
WKYGFVFVGYVS+ETCSYITKYVTKSINGDKVRPR+ISSFGIGSNYL+TEE+ LHKLGNQ
Sbjct 159 WKYGFVFVGYVSDETCSYITKYVTKSINGDKVRPRVISSFGIGSNYLNTEESSLHKLGNQ 218
Query 121 HYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNIVLDRFINPPVEFSWQGQKFSSKLERDEM 180
YQPFMVLNGFQQAMPRYYYNKIFSDVDKQN+V+DR INPPVEFSWQGQKFSSKLERDEM
Sbjct 219 RYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNMVVDRLINPPVEFSWQGQKFSSKLERDEM 278
Query 181 RRSTLNQNITSGLTPALPLPHTERVSSFNRFKKNMDKNKEFK 222
RRSTLNQNI SGLTP LPLPHTERVSSF+ FKK MDKNKEFK
Sbjct 279 RRSTLNQNIASGLTPVLPLPHTERVSSFDIFKKYMDKNKEFK 320
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 78.2 bits (191), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 58/164 (35%), Positives = 89/164 (54%), Gaps = 24/164 (15%)
Query 1 LFLDRLRKDYSK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHH--PLL 57
LF+ RLRK Y++ ++R++ E+G+ GRPHYH ILF P T G H LL
Sbjct 47 LFMKRLRKKYAQYRLRYFLTSEYGSQGGRPHYHMILFGFPFT----------GKHGGDLL 96
Query 58 ASRWKYGFVFVGYVSEETCSYITKYV-TKSINGDKVR------PRIISSF--GIGSNYLD 108
A WK GFV ++ + SY+TKY+ KS+ D ++ P ++ S GIG ++L
Sbjct 97 AECWKNGFVQAHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLR 156
Query 109 TEEAVLHKLGNQHYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNI 152
+ ++L + Y NG + AMPRYY +K++ D K+ +
Sbjct 157 EQILDFYRLHPRDY--VRAFNGMRMAMPRYYADKLYDDDMKEYL 198
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 77.8 bits (190), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 58/164 (35%), Positives = 89/164 (54%), Gaps = 24/164 (15%)
Query 1 LFLDRLRKDYSK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHH--PLL 57
LF+ RLRK Y++ ++R++ E+G+ GRPHYH ILF P T G H LL
Sbjct 81 LFMKRLRKKYAQYRLRYFLTSEYGSQGGRPHYHMILFGFPFT----------GKHGGDLL 130
Query 58 ASRWKYGFVFVGYVSEETCSYITKYV-TKSINGDKVR------PRIISSF--GIGSNYLD 108
A WK GFV ++ + SY+TKY+ KS+ D ++ P ++ S GIG ++L
Sbjct 131 AECWKNGFVQAHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLR 190
Query 109 TEEAVLHKLGNQHYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNI 152
+ ++L + Y NG + AMPRYY +K++ D K+ +
Sbjct 191 EQILDFYRLHPRDY--VRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 76.6 bits (187), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 57/164 (35%), Positives = 88/164 (54%), Gaps = 24/164 (15%)
Query 1 LFLDRLRKDYSK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHH--PLL 57
LF+ RLRK Y + ++R++ E+G+ GRPHYH ILF P T G H LL
Sbjct 81 LFMKRLRKKYDQYRLRYFLTSEYGSQGGRPHYHMILFGFPFT----------GKHGGDLL 130
Query 58 ASRWKYGFVFVGYVSEETCSYITKYV-TKSINGDKVR------PRIISSF--GIGSNYLD 108
A WK GFV ++ + +Y+TKY+ KS+ D ++ P ++ S GIG ++L
Sbjct 131 AECWKNGFVQAHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIPGIGYHFLR 190
Query 109 TEEAVLHKLGNQHYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNI 152
+ ++L + Y NG + AMPRYY +K++ D K+ +
Sbjct 191 EQILDFYRLHPRDY--VRAFNGMRMAMPRYYADKLYDDDMKEYL 232
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 73.9 bits (180), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/157 (32%), Positives = 79/157 (50%), Gaps = 23/157 (15%)
Query 1 LFLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASR 60
LF+ R RK +I+++ V E+G RPHYH I+F V ID +
Sbjct 82 LFMKRARKLQKSKIKYFLVGEYGAQTYRPHYHAIVFGVEN--IDAF-----------LGE 128
Query 61 WKYGFVFVGYVSEETCSYITKYVTKSIN--------GDKVRPRIISSFGIGSNYLDTEEA 112
W+ G V G V+ ++ Y KY TKSI D+ + + S G+G ++L E+
Sbjct 129 WRMGNVHAGTVTAKSIYYTLKYCTKSITEGPDKDPDDDRKPEKALMSKGLGLSHL--TES 186
Query 113 VLHKLGNQHYQPFMVLNGFQQAMPRYYYNKIFSDVDK 149
++ + + F +L G A+PRYY +K+FSD++K
Sbjct 187 MIKYYKDDVSRSFSLLGGTTIALPRYYRDKVFSDIEK 223
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 65.1 bits (157), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 80/156 (51%), Gaps = 20/156 (13%)
Query 1 LFLDRLRKDYSK-QIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLAS 59
LF+ RLRK Y ++R++ E+G GRPHYH ILF P ++ + G LLA
Sbjct 76 LFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFP------FTGKMAGD--LLAE 127
Query 60 RWKYGFVFVGYVSEETCSYITKYV-TKSI------NGDKVRPRIISSF--GIGSNYLDTE 110
W+ GFV ++ + +Y+ KY+ KS+ + K +P ++ S GIG ++ +
Sbjct 128 CWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKAD 187
Query 111 EAVLHKLGNQHYQPFMVLNGFQQAMPRYYYNKIFSD 146
++ + Y G + AMPRYY +K++ D
Sbjct 188 IIEFYRRHPRDY--VRAWAGHKMAMPRYYADKLYDD 221
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 63.2 bits (152), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 49/159 (31%), Positives = 70/159 (44%), Gaps = 23/159 (14%)
Query 2 FLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPL----- 56
F+ RLRK Y +IR + E+G RPHYH +LF D G L
Sbjct 89 FMKRLRKKYPHKIRFFHCGEYGDQNKRPHYHALLFG--HDFKDKKLWSNKGDFKLFVSQE 146
Query 57 LASRWKYGFVFVGYVSEETCSYITKYVTKSINGDKVRP---RIISSFGIGSNYLDTEEAV 113
LA W YGF +G VS +T +Y +YV K + GD + G N + E
Sbjct 147 LAELWPYGFHTIGAVSFDTAAYCARYVMKKVTGDAAASHYREVDLETGEVINEIKPEYCT 206
Query 114 LHKL---GNQHYQP----------FMVLNGFQQAMPRYY 139
+ ++ G + YQ ++V+NG++ PRYY
Sbjct 207 MSRMPGIGYEWYQKYGYHDCHKHDYIVINGYKVRPPRYY 245
>gi|495507506|ref|WP_008232152.1| hypothetical protein [Richelia intracellularis]
gi|471331139|emb|CCH66547.1| hypothetical protein RINTHH_3920 [Richelia intracellularis HH01]
Length=306
Score = 62.8 bits (151), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 49/179 (27%), Positives = 81/179 (45%), Gaps = 43/179 (24%)
Query 2 FLDRLRKDY--------------SKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYS 47
F+ RLRK + S IR++ E+GT GRPHYH LF+ I+
Sbjct 85 FMKRLRKRFKGHKENIDVRTGKSSYPIRYYMAGEYGTHGGRPHYHACLFNFAFEDIEFLR 144
Query 48 PDVPGHH----PLLASRWKYGFVFVGYVSEETCSYITKYVTKSINGD------------- 90
G + L S W +GF VG V+ E+ +Y+ +YV K +N +
Sbjct 145 RTNSGSNLYRSAQLESLWPHGFSSVGDVTFESAAYVARYVMKKMNKEAIEKGQEINWETG 204
Query 91 KVRPRIIS------SFGIGSNYLDTEEAVLHKLGNQHYQPFMVLNGFQQAMPRYYYNKI 143
+V PR+ GIG+N++D ++ + ++++NG + PRYY+ ++
Sbjct 205 EVMPRLPEYNKMSLKPGIGANFIDKYQSDVFP------NDYVIVNGHKAKPPRYYFKRL 257
>gi|575094560|emb|CDL65924.1| unnamed protein product [uncultured bacterium]
Length=320
Score = 62.8 bits (151), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 51/94 (54%), Gaps = 5/94 (5%)
Query 1 LFLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGH----HPL 56
LF RLRK +R+ E+G+ YGRPHYH I+F++P + G +
Sbjct 100 LFWKRLRKR-GVSLRYMACGEYGSTYGRPHYHAIIFNLPPLELKQIGTTSTGFPTFISDV 158
Query 57 LASRWKYGFVFVGYVSEETCSYITKYVTKSINGD 90
++ W GF + VS +TC+Y+ +YVTK I GD
Sbjct 159 ISECWSLGFHTLNPVSFQTCAYVARYVTKKILGD 192
>gi|410493159|ref|YP_006908225.1| replication-associated protein [Dragonfly-associated microphage
1]
gi|406870779|gb|AFS65317.1| replication-associated protein [Dragonfly-associated microphage
1]
Length=270
Score = 58.2 bits (139), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 46/87 (53%), Gaps = 11/87 (13%)
Query 2 FLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASRW 61
FL R R +++R++ V E+G L GRPH+H LF V P ++A W
Sbjct 67 FLKRFRYYLQRKVRYFGVGEYGDLRGRPHFHMALFGVS-----------PLEEGIIAKAW 115
Query 62 KYGFVFVGYVSEETCSYITKYVTKSIN 88
GFV VG +++++ Y+ YVTK +
Sbjct 116 SIGFVHVGDLTKDSAQYLCGYVTKKMT 142
Lambda K H a alpha
0.322 0.140 0.435 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 848296716240