bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_1
Length=326
Score E
Sequences producing significant alignments: (Bits) Value
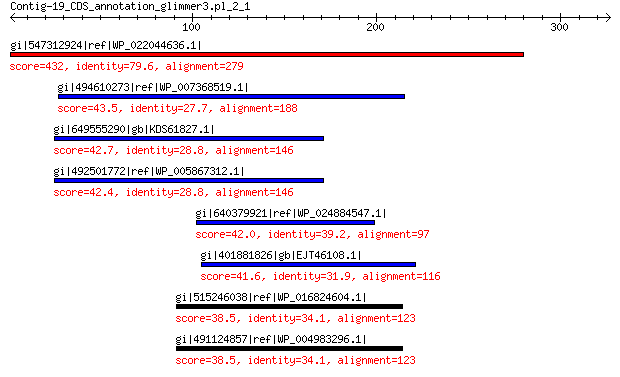
gi|547312924|ref|WP_022044636.1| putative uncharacterized protein 432 2e-148
gi|494610273|ref|WP_007368519.1| hypothetical protein 43.5 0.17
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 42.7 0.20
gi|492501772|ref|WP_005867312.1| hypothetical protein 42.4 0.31
gi|640379921|ref|WP_024884547.1| hypothetical protein 42.0 0.59
gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 41.6 0.84
gi|515246038|ref|WP_016824604.1| hypothetical protein 38.5 7.1
gi|491124857|ref|WP_004983296.1| hypothetical protein 38.5 7.4
>gi|547312924|ref|WP_022044636.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208405|emb|CCZ76640.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=328
Score = 432 bits (1112), Expect = 2e-148, Method: Compositional matrix adjust.
Identities = 222/282 (79%), Positives = 251/282 (89%), Gaps = 3/282 (1%)
Query 1 MSIlaglgaaaaSFALKEGHNAIAQSRNEKNMALEHDYWKRRVNYLEELNKPSRQVAKWR 60
MSILAGLGAAAASFA+KEGHNAIAQSRNEKNMALEHDYWKRRVN LEE+NKPSRQVAKWR
Sbjct 1 MSILAGLGAAAASFAMKEGHNAIAQSRNEKNMALEHDYWKRRVNQLEEMNKPSRQVAKWR 60
Query 61 SAGIAPQAVFGNSPGGAGIATDASAPNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV 120
SAGIAPQAVFGNSPGGAGIATDAS+PNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV
Sbjct 61 SAGIAPQAVFGNSPGGAGIATDASSPNSQTPMGSSDFNFVTTIAERQRMKNEKAIADATV 120
Query 121 NKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRANNEADM 180
+KLNAEA KLRGDTKDP VTKD Q+LEFDWN+VKKQRE+VQL VDEI+KEF+RA NEAD+
Sbjct 121 DKLNAEAGKLRGDTKDPKVTKDLQQLEFDWNIVKKQRERVQLDVDEIDKEFRRAVNEADL 180
Query 181 QIKRGLYSETLAKIDKLIADKDVSEEMKQNLQKQRDLIESQISATQAQTDFTKAQTMTEN 240
QIKRG+YSETL+KIDKLIADK+VSEEMKQNLQKQRDLI++QI +T+AQT +KAQT TE+
Sbjct 181 QIKRGIYSETLSKIDKLIADKEVSEEMKQNLQKQRDLIDAQIDSTKAQTGLSKAQTKTED 240
Query 241 MTRDDRIREIKSRINNLVSSTNLND---ENARSALYSRLADV 279
RD R++ ++ + L+S LND + + + RL D+
Sbjct 241 TLRDGRVKLTGAQTSELLSMAGLNDVRRDREKYETFLRLLDI 282
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 43.5 bits (101), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 52/213 (24%), Positives = 92/213 (43%), Gaps = 27/213 (13%)
Query 27 RNEKNMALEHDYWKRRV-NYLEELNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDASA 85
+N+ M E + + R+ N + + N P+ Q+ ++ AGI P GN G + SA
Sbjct 109 QNQYQMFQEQNAFNERMWNQMNQYNSPAAQMQRYTDAGINPYIAAGNVQTGNAQSALQSA 168
Query 86 PNSQ---------TPMGSSDFNFVTTIAE--RQRMKNEKAIADAT--------VNKLN-A 125
P Q T MG + N I Q +N+ A+A A +++LN A
Sbjct 169 PAPQQHVAQVMPATGMGDAVQNSFAQIGNVISQFAQNQLALAQAKKTDAEASWIDRLNSA 228
Query 126 EAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEINKEFQRANNEADMQIKRG 185
+ KL +T N+ + L D+ + +L D ++ NN D Q ++
Sbjct 229 QMGKLGAETL--NIHNQNSLLGLDYQIKSDTLGNYKLLSDLSVQQAALTNNLVDAQTRKA 286
Query 186 LYSETLAKIDKLI----ADKDVSEEMKQNLQKQ 214
L+ LA ++ I +K V +E+ +++ +Q
Sbjct 287 LFESDLAMVESHIKAKYGEKQVLQEISESVSRQ 319
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 42.7 bits (99), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 42/156 (27%), Positives = 70/156 (45%), Gaps = 16/156 (10%)
Query 25 QSRNEKNMALEHDYWKRRVNYLEELNKPSRQVAKWRSAGIAPQAVFGNS-PGGAGIATDA 83
Q NEK W N E N P++Q+A+ R+AG+ P V+GN G + +T
Sbjct 47 QQENEKAYQRSLKMW----NLQNEYNSPTQQMARIRAAGLNPNLVYGNGVTGNSAGSTPQ 102
Query 84 SAP---NSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKLRGD-----TK 135
P N+ T +N + A Q + A V+ + A+ +R T+
Sbjct 103 YEPAKFNAPTMQAYRGWNLGISDATSQYLAYR--TVKAQVDNMEAQNSLIRQQTATEATR 160
Query 136 DPNVTKDSQRLEFDWNLVKKQRE-QVQLAVDEINKE 170
N+ + R EFD N+ K+ ++ V A+ E+N++
Sbjct 161 QANIAASTSRSEFDLNMAKELKDVSVSSAIAEMNQK 196
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 42.4 bits (98), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 42/156 (27%), Positives = 71/156 (46%), Gaps = 16/156 (10%)
Query 25 QSRNEKNMALEHDYWKRRVNYLEELNKPSRQVAKWRSAGIAPQAVFGNS-PGGAGIATDA 83
Q NEK + W N E N P++Q+A+ R+AG+ P V+GN G + +T
Sbjct 47 QQENEKAYQRSLNMW----NLQNEYNSPTQQMARIRAAGLNPNLVYGNGVTGNSSGSTPQ 102
Query 84 SAP---NSQTPMGSSDFNFVTTIAERQRMKNEKAIADATVNKLNAEAEKLRGD-----TK 135
P N+ T +N + A Q + A V+ + A+ +R TK
Sbjct 103 YEPAKFNAPTMQAYRGWNLGISDAISQFLAYR--TVKAQVDNMEAQNSLIRQQTATEATK 160
Query 136 DPNVTKDSQRLEFDWNLVKKQRE-QVQLAVDEINKE 170
N+ + R EFD N+ K+ ++ V A+ ++N++
Sbjct 161 QANIAASTSRSEFDLNMAKELKDVSVSSAIADMNQK 196
>gi|640379921|ref|WP_024884547.1| hypothetical protein [Streptomyces sp. CNH189]
Length=1293
Score = 42.0 bits (97), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 38/112 (34%), Positives = 59/112 (53%), Gaps = 16/112 (14%)
Query 102 TIAERQRM------KNEKAIADAT--VNKLNAEAEKLRGDTKD--PNVTKDSQRLEFDWN 151
T+AE +R+ K EK IADAT +L AEA + G + + +D++R++ D
Sbjct 945 TVAEAERVRSESVAKAEKLIADATGDAERLRAEAAETVGSAQQHAERIRRDAERVKTDAE 1004
Query 152 -----LVKKQREQVQLAVDEINKEFQRANNEADMQIKRGLYSETLAKIDKLI 198
LV RE+ + +DE KE + +EA Q+ L +ET A+ DKL+
Sbjct 1005 TEAERLVSGAREEAERTLDEARKEANKRRSEAAEQVD-TLITETTAEADKLL 1055
>gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 [Trichosporon asahii var. asahii
CBS 2479]
Length=936
Score = 41.6 bits (96), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 37/121 (31%), Positives = 64/121 (53%), Gaps = 16/121 (13%)
Query 105 ERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAV 164
ER+R+K E A DA V +LNAE E+LR ++P+V + D + V + +EQ+ A+
Sbjct 632 ERRRLKKEVADRDAQVERLNAETERLREQPQEPSVRSEPG----DSSAVAELQEQLDAAM 687
Query 165 DEINK-EFQRANNEADMQIKRGLYSETLAKID----KLIADKDVSEEMKQNLQKQRDLIE 219
+++ E Q + EA+ + + A+++ KL D E+ ++L RD I+
Sbjct 688 SKLSSLEIQHQDLEAE-------HDDATAELEHTRQKLADTNDALEKASEDLYAARDAID 740
Query 220 S 220
S
Sbjct 741 S 741
>gi|515246038|ref|WP_016824604.1| hypothetical protein [Streptomyces viridosporus]
Length=1293
Score = 38.5 bits (88), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 65/138 (47%), Gaps = 22/138 (16%)
Query 91 PMGSSDFNFVTTIAERQRM------KNEKAIADAT--VNKLNAEAEKLRGDTKD--PNVT 140
+ +D T+AE +R+ K EK IADAT +L AEA + G + +
Sbjct 934 TVAETDRLRTETVAEAERVRADSVAKAEKLIADATGDAERLRAEAAQTVGAAQQHAERIR 993
Query 141 KDSQRLEFDWN-----LVKKQREQVQLAVDEINKEFQRANNEADMQIKRGLYSETLAKID 195
+++R+ D LV RE+ + +DE KE + EA Q+ + L +ET A+ D
Sbjct 994 GEAERVRTDAEAEAERLVSSAREESERTLDEARKEANKRRTEAAEQVDK-LITETTAEAD 1052
Query 196 KLIADKDVSEEMKQNLQK 213
KL+ E +Q QK
Sbjct 1053 KLLT------EAQQQAQK 1064
>gi|491124857|ref|WP_004983296.1| hypothetical protein [Streptomyces ghanaensis]
gi|291340080|gb|EFE67036.1| large Ala/Glu-rich protein [Streptomyces ghanaensis ATCC 14672]
Length=1303
Score = 38.5 bits (88), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 65/138 (47%), Gaps = 22/138 (16%)
Query 91 PMGSSDFNFVTTIAERQRM------KNEKAIADAT--VNKLNAEAEKLRGDTKD--PNVT 140
+ +D T+AE +R+ K EK IADAT +L AEA + G + +
Sbjct 944 TVAETDRLRTETVAEAERVRADSVAKAEKLIADATGDAERLRAEAAQTVGAAQQHAERIR 1003
Query 141 KDSQRLEFDWN-----LVKKQREQVQLAVDEINKEFQRANNEADMQIKRGLYSETLAKID 195
+++R+ D LV RE+ + +DE KE + EA Q+ + L +ET A+ D
Sbjct 1004 GEAERVRTDAEAEAERLVSSAREESERTLDEARKEANKRRTEAAEQVDK-LITETTAEAD 1062
Query 196 KLIADKDVSEEMKQNLQK 213
KL+ E +Q QK
Sbjct 1063 KLLT------EAQQQAQK 1074
Lambda K H a alpha
0.311 0.125 0.335 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1835116218150