bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_4
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
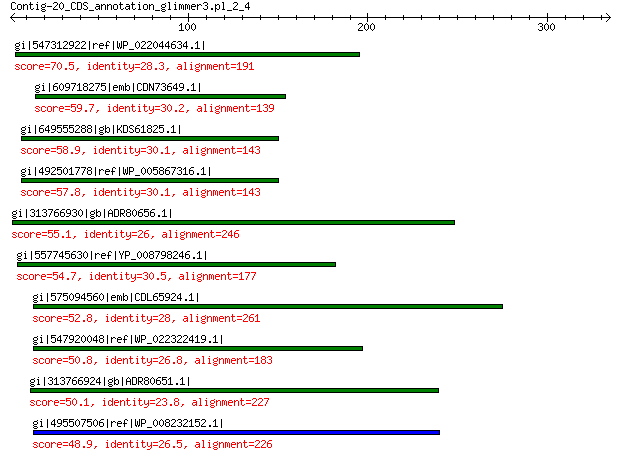
gi|547312922|ref|WP_022044634.1| putative replication initiation... 70.5 2e-10
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 59.7 5e-07
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 58.9 1e-06
gi|492501778|ref|WP_005867316.1| hypothetical protein 57.8 3e-06
gi|313766930|gb|ADR80656.1| putative replication initiation protein 55.1 3e-05
gi|557745630|ref|YP_008798246.1| replication initiator 54.7 3e-05
gi|575094560|emb|CDL65924.1| unnamed protein product 52.8 2e-04
gi|547920048|ref|WP_022322419.1| putative replication protein 50.8 4e-04
gi|313766924|gb|ADR80651.1| putative replication initiation protein 50.1 9e-04
gi|495507506|ref|WP_008232152.1| hypothetical protein 48.9 0.002
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 70.5 bits (171), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 54/205 (26%), Positives = 93/205 (45%), Gaps = 25/205 (12%)
Query 4 PWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSA 63
P DY + VPCG C C + N++ +RL E + K +FVT+T + +
Sbjct 35 PPDYILE---VPCGYCHSCQKSYNNQYRIRLLYELR--KYPPGTCLFVTLTFNDDSLEKF 89
Query 64 LLNPSAFIRMWFERVRRRFGHSIKHAVFQEFG-MHPEQGNEPRLHFHGVLWDVSCS---- 118
+ + +R++ +R R+ +G I+H EFG +H R H+HG+L++V +
Sbjct 90 SKDTNKAVRLFLDRFRKVYGKQIRHWFVCEFGTLH------GRPHYHGILFNVPQALIDG 143
Query 119 -------YNAIREAVKDLGFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
++ + + GFV++ ++D+ Y+ KYV KS+ D+ S I
Sbjct 144 YDSDMPGHHPLLASCWKYGFVFVGYVSDETCSYITKYVTKSINGDKVRPRVISSFGIGSN 203
Query 172 KLKTNLYDF--LQNSRYRRKFISAG 194
L T L N RY+ + G
Sbjct 204 YLNTEESSLHKLGNQRYQPFMVLNG 228
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 59.7 bits (143), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 70/143 (49%), Gaps = 15/143 (10%)
Query 15 PCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAP---EYYDSALLNPS-AF 70
PCG+C EC + + N W+ RL E K KS H FVT+T + Y D+ L++
Sbjct 24 PCGKCLECRKARTNSWFARLTEELKVSKSAH----FVTLTYSDVYLPYSDNGLISLDYRD 79
Query 71 IRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYNAIREAVKDLG 130
+++ +R R+ IK+ + E+G R H+H +++ V + E +G
Sbjct 80 FQLFMKRARKLQKSKIKYFLVGEYG-----AQTYRPHYHAIVFGVENIDAFLGEW--RMG 132
Query 131 FVWISSITDKRLRYVVKYVGKSV 153
V ++T K + Y +KY KS+
Sbjct 133 NVHAGTVTAKSIYYTLKYCTKSI 155
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 58.9 bits (141), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 78/159 (49%), Gaps = 26/159 (16%)
Query 7 YFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLN 66
+ R VPCGRC C + +R W RL+ E + S+FVT+T E+ +A++
Sbjct 8 HLPDRGAVPCGRCVNCRKNKRQSWVYRLQAEA----DEYPFSLFVTLTYDDEHIPTAMIG 63
Query 67 PSAF-----------IRMWFERVRRRFG-HSIKHAVFQEFGMHPEQGNEPRLHFHGVLWD 114
F I+++ +R+R+++ + +++ + E+G QG P H+H +L+
Sbjct 64 EDLFKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG---SQGGRP--HYHMILFG 118
Query 115 VSCS----YNAIREAVKDLGFVWISSITDKRLRYVVKYV 149
+ + + E K+ GFV +T K + YV KY+
Sbjct 119 FPFTGKHGGDLLAECWKN-GFVQAHPLTTKEISYVTKYM 156
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 57.8 bits (138), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 78/159 (49%), Gaps = 26/159 (16%)
Query 7 YFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLN 66
+ R VPCGRC C + +R W RL+ E + S+FVT+T E+ +A++
Sbjct 8 HLPDRGAVPCGRCVNCRKNKRQSWVYRLQAEA----DEYPFSLFVTLTYDDEHMPTAMIG 63
Query 67 PSAF-----------IRMWFERVRRRFG-HSIKHAVFQEFGMHPEQGNEPRLHFHGVLWD 114
F I+++ +R+R+++ + +++ + E+G QG P H+H +L+
Sbjct 64 EDLFKSTVGVVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYG---SQGGRP--HYHMILFG 118
Query 115 VSCS----YNAIREAVKDLGFVWISSITDKRLRYVVKYV 149
+ + + E K+ GFV +T K + YV KY+
Sbjct 119 FPFTGKHGGDLLAECWKN-GFVQAHPLTTKEIAYVTKYM 156
>gi|313766930|gb|ADR80656.1| putative replication initiation protein [Uncultured Microviridae]
Length=402
Score = 55.1 bits (131), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 64/266 (24%), Positives = 114/266 (43%), Gaps = 38/266 (14%)
Query 2 NRPWDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYD 61
N+P+ Y + +PCG+C C Q EW +R E + +H ++ F+T+TI PE +
Sbjct 117 NKPFAY-AKGFNLPCGQCWGCRLQHSREWAIRCMHEAQ----MHDHNCFITLTINPETLE 171
Query 62 SALLNPSAFIRMWFE----RVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGV------ 111
P + + F+ R+RR+ G IK+ E+G R H+H +
Sbjct 172 RR-PRPWSLEKKEFQEFVHRLRRKIGKKIKYFHCGEYG-----DENKRPHYHAIIFGYDF 225
Query 112 ----LWDVSCSYNA-IREAVKDL---GFVWISSITDKRLRYVVKYVGKSVYMDERSADFA 163
LW+ I +++L G+ I + T + YV +YV K + +
Sbjct 226 PDKQLWERKLGNELYISPELENLWPHGYHRIGACTYESAHYVARYVMKRAKGEGPPEQYI 285
Query 164 KSLPITVGKLKTNLYD-FLQNSRYRRKFISAGVGDYLGDFKAPGVASGLWSYTDHKTGCV 222
G+++ +L + + SR +K G+G+ +K + Y H
Sbjct 286 NP---ETGEVEYDLDNQYATMSRGNKKQPQNGIGNQWY-WKYGWTDAHCHDYIVHDG--- 338
Query 223 YRYRIPRYYDKYLSQ-DALLFRKIST 247
+ ++PRYYDK L + D F+++
Sbjct 339 IKMKVPRYYDKELEKYDPEYFQELKA 364
>gi|557745630|ref|YP_008798246.1| replication initiator [Marine gokushovirus]
gi|530695343|gb|AGT39900.1| replication initiator [Marine gokushovirus]
Length=312
Score = 54.7 bits (130), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 54/210 (26%), Positives = 91/210 (43%), Gaps = 44/210 (21%)
Query 5 WDYFTQRIMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSAL 64
W + PCG C+ C +++ EW VR E + ++ +S F+T+T YD+
Sbjct 24 WPEMYPPMTRPCGYCKGCRFKKQQEWTVRCLNEQQILETKGRSSSFITLT-----YDNKH 78
Query 65 LNPSAFI--RMWFERVR----RRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLW----- 113
L P+ + W + +R R G SI++ E+G N R HFH +L+
Sbjct 79 LPPNNSLDYTHWQKFIRSLKKRNNGKSIRYFGVGEYGE-----NFGRPHFHAILFGHTFN 133
Query 114 DVSCSYNAIREAVKDL------------GFVWISSITDKRLRYVVKYVGKSV-------- 153
D+ ++ I ++ + L GFV + +T + + YV YV K +
Sbjct 134 DLIPMHSNISKSQQLLSAWCEPLTQEPRGFVSVGDVTPESISYVCGYVQKKIFGQGQAAH 193
Query 154 --YMDERSADFAKSLPITVGKLKTNLYDFL 181
Y+D+ S + P T GK+ L F+
Sbjct 194 YKYIDKDSGEITTKKP-TNGKIYRLLKSFM 222
>gi|575094560|emb|CDL65924.1| unnamed protein product [uncultured bacterium]
Length=320
Score = 52.8 bits (125), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 73/299 (24%), Positives = 126/299 (42%), Gaps = 73/299 (24%)
Query 14 VPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSV------FVTITIAPEYYDS-ALLN 66
+PCG+C C RL+R HH S+ F+T+T +PE+ L
Sbjct 45 IPCGQCIGC----------RLDRSLDSAVRAHHESLLYDRNYFLTLTYSPEHLPPFGSLI 94
Query 67 PSAFIRMWFERVRRRFGHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWD--------VSCS 118
P W +R+R+R G S+++ E+G R H+H ++++ + +
Sbjct 95 PRDLTLFW-KRLRKR-GVSLRYMACGEYG-----STYGRPHYHAIIFNLPPLELKQIGTT 147
Query 119 YNAIREAVKD-------LGFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLPITVG 171
+ D LGF ++ ++ + YV +YV K + D + + K P+T G
Sbjct 148 STGFPTFISDVISECWSLGFHTLNPVSFQTCAYVARYVTKKILGDGKQV-YEKFDPVT-G 205
Query 172 KLKTNLYDFLQNSRYRRKFISAGVG-DYLGDFKAPGVASGLWSYTDHKTGCVY----RYR 226
++ + +F SR+ K G+G DY + W +K C +++
Sbjct 206 EVDCRVKEF---SRWSTK---PGIGHDYFMKY---------WR-DFYKIDCCLINNKKFK 249
Query 227 IPRYYDKYLSQD------ALLFRKISTAWTYASAFGGSMALGFLREV-----AERVLRP 274
IPRYYD+ L +D + ++I +A Y + +RE AER+LRP
Sbjct 250 IPRYYDRLLLRDHPDVFEIVKQKRILSAQDYRLTPDAQKSRLLVREEVKRLRAERLLRP 308
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 50.8 bits (120), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 49/207 (24%), Positives = 90/207 (43%), Gaps = 35/207 (17%)
Query 14 VPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLNPSAF--- 70
VPCG C C + +R W RL+ E K + S+FVT+T E+ + F
Sbjct 10 VPCGWCVNCRQNKRQSWVYRLQAEAKE----YPLSLFVTLTYDDEHLPIERIGSDLFQTN 65
Query 71 --------IRMWFERVRRRF-GHSIKHAVFQEFGMHPEQGNEPRLHFHGVLWDVSCSYNA 121
++++ +R+R+++ + +++ V E+G R H+H +L+ +
Sbjct 66 VAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYG-----AKNGRPHYHMILFGFPFTGKM 120
Query 122 IREAVKDL---GFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLP---------IT 169
+ + + GFV +T K + YV KY+ + E D K P I
Sbjct 121 AGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIG 180
Query 170 VGKLKTNLYDFLQNSRYRRKFISAGVG 196
G +K ++ +F + R+ R ++ A G
Sbjct 181 FGFMKADIIEFYR--RHPRDYVRAWAG 205
>gi|313766924|gb|ADR80651.1| putative replication initiation protein [Uncultured Microviridae]
Length=285
Score = 50.1 bits (118), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 54/259 (21%), Positives = 100/259 (39%), Gaps = 52/259 (20%)
Query 12 IMVPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLNPSAFI 71
+ V C +C C W R+E E+ + N F+T+T E+ +
Sbjct 1 MEVACSQCIGCRLDHAGMWASRIEHESSLYDDSNGN-CFITLTYDEEHLPQDWSLDKSHF 59
Query 72 RMWFERVRRRFGHSIKHAVFQEFGMHPEQG---------NEPRLHFHGVLWDVSCSYNAI 122
+ + +R+R+R+ I++ E+G + G N R H+H +L+++ +
Sbjct 60 QKFMKRLRKRYPQKIRYYHCGEYGENCRHGIHTTLCPGCNVGRPHYHAILFNIDFHDRVL 119
Query 123 REAVKDL--------------GFVWISSITDKRLRYVVKYVGKSVYMDERSADFAKSLPI 168
K + GF + +T + YV +Y K V ++ D +S+ +
Sbjct 120 VGQSKGIPHFTSDTLTEIWGHGFTQVGDLTAQSAGYVARYALKKV-TGTQAEDHYRSIDL 178
Query 169 TVGKLKTNLYDFLQNSRYRRKFISAGVG---------DYLGDFKAPGVASGLWSYTDHKT 219
T G++ ++ SR G+G D + P V G+ K
Sbjct 179 TTGEVTYVRPEYATMSR------KPGIGKEWYEKYKKDMYPSNQTPSVGGGV------KN 226
Query 220 GCVYRYRIPRYYDKYLSQD 238
G IPR+YDK + ++
Sbjct 227 G------IPRFYDKLMEKE 239
>gi|495507506|ref|WP_008232152.1| hypothetical protein [Richelia intracellularis]
gi|471331139|emb|CCH66547.1| hypothetical protein RINTHH_3920 [Richelia intracellularis HH01]
Length=306
Score = 48.9 bits (115), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 60/258 (23%), Positives = 108/258 (42%), Gaps = 59/258 (23%)
Query 14 VPCGRCEECLRQQRNEWYVRLERETKYQKSLHHNSVFVTITIAPEYYDSALLNP--SAFI 71
+PCG CE CL ++ +W VR E + L + FVT+T Y ++ N +
Sbjct 31 LPCGHCEGCLLERSRQWAVRCMHEAQ----LWERNCFVTLT----YEETPPWNSLRHSDF 82
Query 72 RMWFERVRRRF-GHS-------------IKHAVFQEFGMHPEQGNEPRLHFHGVLWD--- 114
+ + +R+R+RF GH I++ + E+G H G P H+H L++
Sbjct 83 QKFMKRLRKRFKGHKENIDVRTGKSSYPIRYYMAGEYGTH---GGRP--HYHACLFNFAF 137
Query 115 --------VSCSYNAIR----EAVKDLGFVWISSITDKRLRYVVKYVGKSVYMDERSADF 162
+ N R E++ GF + +T + YV +YV K M++ + +
Sbjct 138 EDIEFLRRTNSGSNLYRSAQLESLWPHGFSSVGDVTFESAAYVARYVMKK--MNKEAIEK 195
Query 163 AKSLPITVGKLKTNLYDFLQNSRYRRKFISAGVG-DYLGDFKAPGVASGLWSYTDHKTGC 221
+ + G++ L Y + + G+G +++ +++ + HK
Sbjct 196 GQEINWETGEVMPRL------PEYNKMSLKPGIGANFIDKYQSDVFPNDYVIVNGHKA-- 247
Query 222 VYRYRIPRYYDKYLSQDA 239
+ PRYY K L Q A
Sbjct 248 ----KPPRYYFKRLKQAA 261
Lambda K H a alpha
0.326 0.140 0.444 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1917593351550