bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-20_CDS_annotation_glimmer3.pl_2_5
Length=648
Score E
Sequences producing significant alignments: (Bits) Value
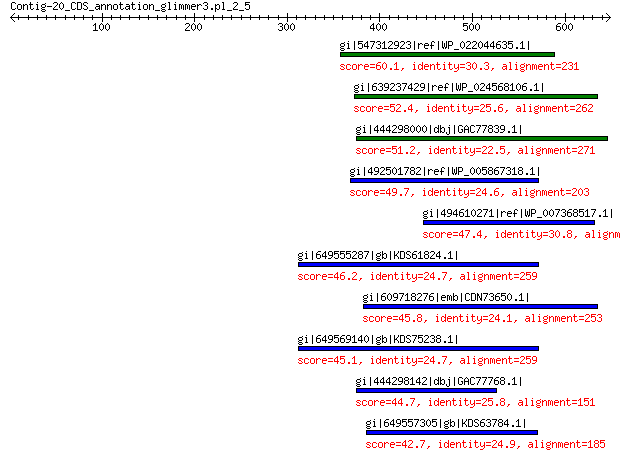
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 60.1 2e-06
gi|639237429|ref|WP_024568106.1| hypothetical protein 52.4 0.001
gi|444298000|dbj|GAC77839.1| major capsid protein 51.2 0.002
gi|492501782|ref|WP_005867318.1| hypothetical protein 49.7 0.006
gi|494610271|ref|WP_007368517.1| capsid protein 47.4 0.029
gi|649555287|gb|KDS61824.1| capsid family protein 46.2 0.078
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 45.8 0.088
gi|649569140|gb|KDS75238.1| capsid family protein 45.1 0.13
gi|444298142|dbj|GAC77768.1| major capsid protein 44.7 0.15
gi|649557305|gb|KDS63784.1| capsid family protein 42.7 0.46
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 60.1 bits (144), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 70/269 (26%), Positives = 106/269 (39%), Gaps = 48/269 (18%)
Query 357 CPSSPDRFSRLMPPGDSNS---------DVDF-TGVKT-IPQLAVATRLQEYKDLIGASG 405
P SPD F ++ G S + D++ TG +P+L + T++Q + D + SG
Sbjct 15 VPYSPDLFGNIIKQGSSPAVEIEVMNALDLNISTGFSVAVPELRLRTKIQNWMDRLFVSG 74
Query 406 SRYSDWLYTFFASKIEHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSI 465
R D T + +K + K F + +A +G A GE A LGQ+ +
Sbjct 75 GRVGDVFRTLWGTKSSAIYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACV 134
Query 466 ----AFNTVLGREQTYYFKEPG--YIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYND 519
F+ G + YY KEPG + ML P Y G+ PD D FNP N
Sbjct 135 DRYCDFSGHSGID--YYAKEPGTFMLITMLVPEPAYS-QGLHPDLASISFGDDFNPELNG 191
Query 520 IGYQDVPFWRI-----GYGWKAASESQS----------------MTVAKEPCYNEFRSSY 558
IG+Q VP R G+ + + S ++V +E ++ R+ Y
Sbjct 192 IGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEVAWSWLRTDY 251
Query 559 DEVLGSLQSTLTPKASVPLQSYWVQQRDF 587
+ G + YWV R F
Sbjct 252 SRLHGDFAQNGNYQ-------YWVLTRRF 273
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 52.4 bits (124), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 67/273 (25%), Positives = 112/273 (41%), Gaps = 36/273 (13%)
Query 373 SNSDVDFTGVK--TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIE--HVDRPKL 428
SN VD TI L A +LQE+ + +GSRY++ + +FF K + RP+
Sbjct 286 SNLGVDLKTASGSTINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEF 345
Query 429 LFsssvmvnsqvvmnqAGQ-----SGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPG 483
L + + V+ Q+ G G ++G+ GG F F+E G
Sbjct 346 LGGNKTPILISEVLQQSSTDSTTPQGNMAGHGISVGKEGGFSKF-----------FEEHG 394
Query 484 YIFDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQS 542
Y+ ++++ P + GI + ++ DYF P + IG Q V I +
Sbjct 395 YVIGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQPVYNKEI-FAKNVGDYDSG 453
Query 543 MTVAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYMIGLSSNPNEISPS 602
P Y+E++ S + G + TL +W R F SS P +++
Sbjct 454 GVFGYVPRYSEYKYSPSTIHGDFKDTLY---------FWHLGRIFD----SSAPPKLNRD 500
Query 603 MLFTNLSTVNNPFA-SDMEDNFFVNMSYKVVVK 634
+ N S ++ FA D D F+ ++ K+ K
Sbjct 501 FIEVNKSGLSRIFAVEDNSDKFYCHLYQKITAK 533
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 51.2 bits (121), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 61/278 (22%), Positives = 111/278 (40%), Gaps = 33/278 (12%)
Query 375 SDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWL-YTFFASKIEHVDRPKLLFsss 433
+D+ TI + A +Q Y++ GSRY+++L Y K + RP+ + +
Sbjct 228 ADLQAATGGTINDIRRAFAIQRYQEARSRYGSRYTEYLRYLGVNPKDARLQRPEYMGGGT 287
Query 434 vmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTY--YFKEPGYIFDMLTI 491
+N V+ + + G + + +G R Y Y +E GYI ML++
Sbjct 288 TQINFSEVLQTSPE--IPGEDQVSQFGVGDMYGHGIAAMRSNKYRRYIEEHGYIISMLSV 345
Query 492 RPVYFWT-GIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPC 550
RP +T GI +L DY+ IG Q++ I A +E T
Sbjct 346 RPKTMYTNGIHRSWLRLTKEDYYQKELEHIGQQEIMNNEIYADEGAGTE----TFGYNDR 401
Query 551 YNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYMIGLSSNPNEISPSML---FTN 607
Y+E+R + V + L +YW R+F +P +L F +
Sbjct 402 YSEYRETPSHVSAEFRGIL---------NYWHMAREFE-----------APPVLNQSFVD 441
Query 608 LSTVNNPFASDMEDNFFVNMSYKVVVKNLVNKSFATRL 645
+D ++ + +K+V + L++++ A R+
Sbjct 442 CDATKRIHNEQTQDALWIMIQHKMVARRLLSRNAAPRI 479
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 49.7 bits (117), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 50/206 (24%), Positives = 90/206 (44%), Gaps = 12/206 (6%)
Query 368 MPPGDSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDR 425
+ P + +VD GV +I L + LQ + + SGSRY + + + F S + R
Sbjct 282 LEPDNFQVNVDELGV-SINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQR 340
Query 426 PKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYI 485
P+ L ++ V+ + + A + G S N R YF+E GYI
Sbjct 341 PQFLGGGRTPISVSEVLQTSATD--STSPQANMAGHGISAGVNHGFKR----YFEEHGYI 394
Query 486 FDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMT 544
+++IRP + G+ D+ ++ D++ P + +G Q++ + AS + T
Sbjct 395 IGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNNG--T 452
Query 545 VAKEPCYNEFRSSYDEVLGSLQSTLT 570
P Y E++ S +EV G + +
Sbjct 453 FGYTPRYAEYKYSMNEVHGDFRGNMA 478
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 47.4 bits (111), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 57/216 (26%), Positives = 92/216 (43%), Gaps = 39/216 (18%)
Query 447 QSGFAGG--EAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRPVYFWTG--IRP 502
QS F G E+ LG +GG ++ + KE G I + ++ P + G P
Sbjct 310 QSEFDRGADESPCLGDLGGK-GVGSLNSSSIDFDVKEHGIIMCIYSVVPQTEYNGTYFDP 368
Query 503 DYLEYRGPDYFNPIYNDIGYQ------------DVPF-------WRIGYGWKAAS-ESQS 542
+ R D+F P + D+GYQ D P R+ G+ +S E+ +
Sbjct 369 FNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDNPVPDGPEKQKRLAAGYPLSSIEANN 428
Query 543 MTVAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQR-DFYMIGLSSNPNEI-- 599
+ + YNE+++S D V G +S L+ SYW R DF G + + +
Sbjct 429 RLLGWQVRYNEYKTSRDLVFGEFESGLS-------LSYWCSPRYDFGFDGKAGDKKLVNS 481
Query 600 --SPSMLFTNLSTVNNPF--ASDMEDNFFVNMSYKV 631
SP+ + N S +N F ++ D+F VN + V
Sbjct 482 PWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDV 517
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 46.2 bits (108), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 64/275 (23%), Positives = 111/275 (40%), Gaps = 25/275 (9%)
Query 312 AYPYDVQEPKVNWNNGSGTDTGTPSKVYFAATLNVPFLAAHPMA---VCPSSPDRFS--- 365
A P+ + P+V G + K FAA F P++ V S+P S
Sbjct 216 ALPWVQRGPEVTVPINGGGEIPVEMKEGFAAQKITTFPDRKPISGSEVLYSAPSVLSYGQ 275
Query 366 -------RLMPPGDSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA- 417
L+ P + + D GV I + + LQ + + SGSRY + + + F
Sbjct 276 IGSIKGQALIEPDNFVVNTDQMGV-NINDIRTSNALQRWFERNARSGSRYIEQILSHFGV 334
Query 418 -SKIEHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQT 476
S + RP+ L ++ V+ + S + A + G S N R
Sbjct 335 RSSDARLQRPQFLGGGRTPISVSEVLQTS--STDSTSPQANMAGHGISAGVNHGFTR--- 389
Query 477 YYFKEPGYIFDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWK 535
YF+E GYI +++IRP + G+ D+ ++ D++ P + +G Q++ +
Sbjct 390 -YFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNES 448
Query 536 AASESQSMTVAKEPCYNEFRSSYDEVLGSLQSTLT 570
A+ T P Y E++ S +EV G + +
Sbjct 449 DAANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMA 481
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 45.8 bits (107), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 61/262 (23%), Positives = 111/262 (42%), Gaps = 34/262 (13%)
Query 382 VKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIE--HVDRPKLLFsssvmvnsq 439
V T+ L A +LQE+ + +GSRY++ + +FF K + RP+ L + +
Sbjct 288 VSTVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMIS 347
Query 440 vvmnqAGQ-----SGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP- 493
V+ Q+ G G +G+ GG + +F+E GY+ ++++ P
Sbjct 348 EVLQQSATDSTTPQGNMAGHGIGIGKDGGF-----------SRFFEEHGYVIGLMSVIPK 396
Query 494 VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWKAASESQSMTVAKEPCYNE 553
+ GI + + DYF P + IG Q V I A +S+++ P Y+E
Sbjct 397 TSYSQGIPRHFSKSDKFDYFWPQFEHIGEQPVYNKEIFAKNIDAFDSEAV-FGYLPRYSE 455
Query 554 FRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYMIGLSSNPNEISPSMLFTNLSTVNN 613
++ S V G + L +W R F + P ++ S + + + ++
Sbjct 456 YKFSPSTVHGDFKDDLY---------FWHLGRIFD----TDKPPVLNQSFIECDKNALSR 502
Query 614 PFA-SDMEDNFFVNMSYKVVVK 634
FA D D F+ ++ K+ K
Sbjct 503 IFAVEDDTDKFYCHLYQKITAK 524
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 45.1 bits (105), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 64/275 (23%), Positives = 111/275 (40%), Gaps = 25/275 (9%)
Query 312 AYPYDVQEPKVNWNNGSGTDTGTPSKVYFAATLNVPFLAAHPMA---VCPSSPDRFS--- 365
A P+ + P+V G + K FAA F P++ V S+P S
Sbjct 65 ALPWVQRGPEVTVPINGGGEIPVEMKEGFAAQKITTFPDRKPISGSEVLYSAPSVLSYGQ 124
Query 366 -------RLMPPGDSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA- 417
L+ P + + D GV I + + LQ + + SGSRY + + + F
Sbjct 125 IGSIKGQALIEPDNFVVNTDQMGV-NINDIRTSNALQRWFERNARSGSRYIEQILSHFGV 183
Query 418 -SKIEHVDRPKLLFsssvmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQT 476
S + RP+ L ++ V+ + S + A + G S N R
Sbjct 184 RSSDARLQRPQFLGGGRTPISVSEVLQTS--STDSTSPQANMAGHGISAGVNHGFTR--- 238
Query 477 YYFKEPGYIFDMLTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWK 535
YF+E GYI +++IRP + G+ D+ ++ D++ P + +G Q++ +
Sbjct 239 -YFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNES 297
Query 536 AASESQSMTVAKEPCYNEFRSSYDEVLGSLQSTLT 570
A + T P Y E++ S +EV G + +
Sbjct 298 DA--ANEGTFGYTPRYAEYKYSQNEVHGDFRGNMA 330
>gi|444298142|dbj|GAC77768.1| major capsid protein [uncultured marine virus]
Length=299
Score = 44.7 bits (104), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 70/153 (46%), Gaps = 6/153 (4%)
Query 375 SDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWL-YTFFASKIEHVDRPKLLFsss 433
+D+ G I L A LQ Y++ G+R++++L Y +S + RP+++ +
Sbjct 111 ADLSQAGAININDLREAFALQRYQEARNLYGARFTEYLRYLGISSSXGRLQRPEMISTGK 170
Query 434 vmvnsqvvmnqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP 493
+N V+N G SG + LG+MGG V Y+ +E G+I ++++RP
Sbjct 171 SNINFSEVLNTTGPSGV---DDHPLGEMGGH-GIAGVKSNRARYFCEEHGHIISLMSVRP 226
Query 494 -VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDV 525
+ T + DY+ IG ++V
Sbjct 227 KTIYMTTQHKQFDRESKEDYWQKELQAIGMEEV 259
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 42.7 bits (99), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 46/189 (24%), Positives = 83/189 (44%), Gaps = 13/189 (7%)
Query 385 IPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLLFsssvmvnsqvvm 442
I + + LQ + + SGSRY + + + F S + RP+ L ++ V+
Sbjct 5 INDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVL 64
Query 443 nqAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDMLTIRP-VYFWTGIR 501
+ S + A + G S N R YF+E GYI +++IRP + G+
Sbjct 65 QTS--STDSTSPQANMAGHGISAGVNHGFTR----YFEEHGYIMGIMSIRPRTGYQQGVP 118
Query 502 PDYLEYRGPDYFNPIYNDIGYQDVPFWRIGYGWK-AASESQSMTVAKEPCYNEFRSSYDE 560
D+ ++ D++ P + +G Q++ + AA+E T P Y E++ S +E
Sbjct 119 KDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEG---TFGYTPRYAEYKYSQNE 175
Query 561 VLGSLQSTL 569
V G + +
Sbjct 176 VHGDFRGNM 184
Lambda K H a alpha
0.320 0.136 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4913515649712