bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_2
Length=341
Score E
Sequences producing significant alignments: (Bits) Value
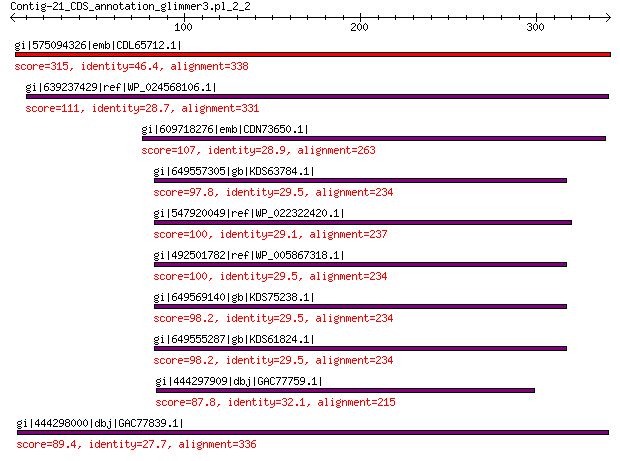
gi|575094326|emb|CDL65712.1| unnamed protein product 315 7e-97
gi|639237429|ref|WP_024568106.1| hypothetical protein 111 6e-24
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 107 2e-22
gi|649557305|gb|KDS63784.1| capsid family protein 97.8 2e-20
gi|547920049|ref|WP_022322420.1| capsid protein VP1 100 5e-20
gi|492501782|ref|WP_005867318.1| hypothetical protein 100 5e-20
gi|649569140|gb|KDS75238.1| capsid family protein 98.2 8e-20
gi|649555287|gb|KDS61824.1| capsid family protein 98.2 3e-19
gi|444297909|dbj|GAC77759.1| major capsid protein 87.8 6e-17
gi|444298000|dbj|GAC77839.1| major capsid protein 89.4 2e-16
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 315 bits (806), Expect = 7e-97, Method: Compositional matrix adjust.
Identities = 157/338 (46%), Positives = 222/338 (66%), Gaps = 14/338 (4%)
Query 4 FVGLTVGDVVTRADDGTYSIQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 63
VGLT ++ + D G T +VDE+G+ Y V ++ +GE L GV+Y P+
Sbjct 435 LVGLTTYEIRSVNDAGHEVTTVNTAIVDEEGNAYKVDFE--SNGEALKGVNYTPLKAGEA 492
Query 64 VTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 123
V + +L + SG +I R VNAYQ++LELN +GFSYK+I++GR+D+++R+
Sbjct 493 V-------NMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRY 545
Query 124 DELLMPEFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVF 183
D L MPE++GGI+R++ + + QT++ G+ G Y +LGS++G+A +G+T +I VF
Sbjct 546 DALNMPEYLGGITRDIVVNPITQTVETTGS---GSYVGSLGSQSGLATCFGNTDGSISVF 602
Query 184 CDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVG 243
CDEES ++G++ V P+PVY LLPK Y LD + PEFD IG+QPI KE+ PM V
Sbjct 603 CDEESIVMGIMYVMPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYAKELGPMQCVQ 662
Query 244 DDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSD 303
DD + FGYQRPWYEYVAK D AHGLF ++NFIM R F +P+LGQ F ++
Sbjct 663 DDIDP--NTVFGYQRPWYEYVAKPDRAHGLFLSSLRNFIMFRSFDNVPELGQSFTVMQPG 720
Query 304 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 341
+VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 721 SVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 758
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 111 bits (278), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 95/333 (29%), Positives = 146/333 (44%), Gaps = 26/333 (8%)
Query 10 GDVVTRADDGTYSIQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTPVTAINS 69
G+ + DG+ S T EDGS V DG + V+ PV NS
Sbjct 236 GNTFVKKPDGSLS---HTGFRLEDGS-------VPADGIGHLMVETSSTGNSNPVNIDNS 285
Query 70 YAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMP 129
L T GS TI LR Q++LE N R G Y + + + + L P
Sbjct 286 SNLGVDLKTASGS--TINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRP 343
Query 130 EFIGGISRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESY 189
EF+GG + + V Q QG A G G G + F +E Y
Sbjct 344 EFLGGNKTPILISEVLQQSSTDSTTPQGNMA-GHGISVGKEGGFSK-------FFEEHGY 395
Query 190 IIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQ 249
+IGL++V P Y+Q +P+ F D++ P+F+ IG QP+ KEI N VGD D+
Sbjct 396 VIGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKN-VGDYDS-- 452
Query 250 LSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVF--KGLPQLGQQFLLVDSDTVNQ 307
FGY + EY + HG F+ + + + R+F P+L + F+ V+ +++
Sbjct 453 -GGVFGYVPRYSEYKYSPSTIHGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNKSGLSR 511
Query 308 VFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 340
+F+V + +DK + ++ TA+ +S P
Sbjct 512 IFAVEDNSDKFYCHLYQKITAKRKMSYFGDPSF 544
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 107 bits (267), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 76/265 (29%), Positives = 124/265 (47%), Gaps = 16/265 (6%)
Query 76 LATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGI 135
+A+E S T+ LR Q++LE N R G Y + + + + L PEF+GG
Sbjct 283 MASENVS--TVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGN 340
Query 136 SRELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLT 195
+ + + + Q DS G GI G + F +E Y+IGL++
Sbjct 341 KSPI---MISEVLQQSATDSTTPQGNMAGHGIGIGKDGGFSR-----FFEEHGYVIGLMS 392
Query 196 VTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFG 255
V P Y+Q +P+ F + D++ P+F+ IG QP+ KEI NI D FG
Sbjct 393 VIPKTSYSQGIPRHFSKSDKFDYFWPQFEHIGEQPVYNKEIFAKNI----DAFDSEAVFG 448
Query 256 YQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVF--KGLPQLGQQFLLVDSDTVNQVFSVTE 313
Y + EY + HG F+ D+ + + R+F P L Q F+ D + ++++F+V +
Sbjct 449 YLPRYSEYKFSPSTVHGDFKDDLYFWHLGRIFDTDKPPVLNQSFIECDKNALSRIFAVED 508
Query 314 YTDKIFGYVKFNATARLPISRVAIP 338
TDK + ++ TA+ +S P
Sbjct 509 DTDKFYCHLYQKITAKRKMSYFGDP 533
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 69/234 (29%), Positives = 109/234 (47%), Gaps = 15/234 (6%)
Query 83 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 142
G I +R NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 2 GVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 61
Query 143 TVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVY 202
V QT DS A G G+ ++ + +E YI+G++++ P Y
Sbjct 62 EVLQT---SSTDSTSPQANMAGH-----GISAGVNHGFTRYFEEHGYIMGIMSIRPRTGY 113
Query 203 TQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYE 262
Q +PKDF +D Y PEF +G Q I +E+ + + D TFGY + E
Sbjct 114 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL----YLNESDAAN-EGTFGYTPRYAE 168
Query 263 YVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTD 316
Y + HG FR +M + ++R+FK P L F V+ + N+VF+ E +D
Sbjct 169 YKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTF--VECNPSNRVFATAETSD 220
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 100 bits (249), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 69/237 (29%), Positives = 108/237 (46%), Gaps = 15/237 (6%)
Query 83 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 142
G I LR NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 310 GININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVS 369
Query 143 TVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVY 202
V QT Q A G+ +N + + +E YIIG++++TP Y
Sbjct 370 EVLQTSSTDETSPQANMAGH--------GISAGINNGFKHYFEEHGYIIGIMSITPRSGY 421
Query 203 TQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYE 262
Q +P+DF +D Y PEF + Q I +E + +D + TFGY + E
Sbjct 422 QQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQE-----LFVSEDAAYNNGTFGYTPRYAE 476
Query 263 YVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTDKIF 319
Y AHG FR ++ + ++R+F+ P L F V+ N+VF+ +E D F
Sbjct 477 YKYHPSEAHGDFRGNLSFWHLNRIFEDKPNLNTTF--VECKPSNRVFATSETEDDKF 531
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 100 bits (248), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 69/234 (29%), Positives = 109/234 (47%), Gaps = 15/234 (6%)
Query 83 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 142
G +I LR NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 295 GVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 354
Query 143 TVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVY 202
V QT DS A G G+ ++ + + +E YIIG++++ P Y
Sbjct 355 EVLQT---SATDSTSPQANMAGH-----GISAGVNHGFKRYFEEHGYIIGIMSIRPRTGY 406
Query 203 TQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYE 262
Q +PKDF +D Y PEF +G Q I +E+ ++ TFGY + E
Sbjct 407 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNN-----GTFGYTPRYAE 461
Query 263 YVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTD 316
Y + HG FR +M + ++R+F P L F V+ + N+VF+ E +D
Sbjct 462 YKYSMNEVHGDFRGNMAFWHLNRIFSESPNLNTTF--VECNPSNRVFATAETSD 513
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 98.2 bits (243), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 69/234 (29%), Positives = 109/234 (47%), Gaps = 15/234 (6%)
Query 83 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 142
G I +R NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 147 GVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 206
Query 143 TVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVY 202
V QT DS A G G+ ++ + +E YI+G++++ P Y
Sbjct 207 EVLQT---SSTDSTSPQANMAGH-----GISAGVNHGFTRYFEEHGYIMGIMSIRPRTGY 258
Query 203 TQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYE 262
Q +PKDF +D Y PEF +G Q I +E+ + + D TFGY + E
Sbjct 259 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL----YLNESDAAN-EGTFGYTPRYAE 313
Query 263 YVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTD 316
Y + HG FR +M + ++R+FK P L F V+ + N+VF+ E +D
Sbjct 314 YKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTF--VECNPSNRVFATAETSD 365
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 98.2 bits (243), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 69/234 (29%), Positives = 109/234 (47%), Gaps = 15/234 (6%)
Query 83 GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMR 142
G I +R NA Q++ E N R G Y + + + + L P+F+GG +S+
Sbjct 298 GVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 357
Query 143 TVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPVY 202
V QT DS A G G+ ++ + +E YI+G++++ P Y
Sbjct 358 EVLQT---SSTDSTSPQANMAGH-----GISAGVNHGFTRYFEEHGYIMGIMSIRPRTGY 409
Query 203 TQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQRPWYE 262
Q +PKDF +D Y PEF +G Q I +E+ + + D TFGY + E
Sbjct 410 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL----YLNESDAAN-EGTFGYTPRYAE 464
Query 263 YVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFLLVDSDTVNQVFSVTEYTD 316
Y + HG FR +M + ++R+FK P L F V+ + N+VF+ E +D
Sbjct 465 YKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTF--VECNPSNRVFATAETSD 516
>gi|444297909|dbj|GAC77759.1| major capsid protein, partial [uncultured marine virus]
Length=257
Score = 87.8 bits (216), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 69/221 (31%), Positives = 102/221 (46%), Gaps = 22/221 (10%)
Query 84 FTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGG------ISR 137
T+ LR Q++LE+N R G Y + + W + L PE++GG +S
Sbjct 43 LTVNDLRLAIRIQEWLEVNARAGSRYVEHLLAHWGVRSSDARLDRPEYLGGGKQPVLVSE 102
Query 138 ELSMRTVEQTIDQQGADSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVT 197
LS T E IDQ+ + Q A G + G SN + +E +I+G+++V
Sbjct 103 VLS--TAEVAIDQEISIPQANMA---GHGISVGG-----SNRFKKRFEEHGHILGIMSVI 152
Query 198 PVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICPMNIVGDDDTEQLSKTFGYQ 257
P Y Q + + F D+Y PEF +G Q + E+ +G DDTE TFGYQ
Sbjct 153 PRTAYQQGVDRSFSREDKFDYYFPEFAHLGEQSVNNYEV----YMG-DDTEN-HDTFGYQ 206
Query 258 RPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFL 298
+ EY K G FR ++ + M R F P LG +F+
Sbjct 207 SRYAEYKYKNSMVTGDFRDNLDFWHMGRQFATRPVLGDEFV 247
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 89.4 bits (220), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 93/342 (27%), Positives = 149/342 (44%), Gaps = 29/342 (8%)
Query 5 VGLTVGDVVTRADDGTY-SIQKQTVLVDEDGS---KYGVSYKVSEDGERLVGVDYDP-VS 59
V L +GD GT S Q + V+E G +YG ++ + D DP
Sbjct 161 VTLPLGDRAPIYGIGTTGSPATQNINVNETGGVNREYGAAWSSETTNAIVAEHDPDPGAG 220
Query 60 EKTPVTAINSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDI 119
P YA+L A +G TI +R A Q++ E R G Y + ++ +
Sbjct 221 SDDP----GIYADLQA-----ATGGTINDIRRAFAIQRYQEARSRYGSRYTEYLR-YLGV 270
Query 120 DIRFDELLMPEFIGGISRELSMRTVEQTIDQ-QGADSQGQYAEALGSKTGIAGVYGSTSN 178
+ + L PE++GG + +++ V QT + G D Q+ +G G G+ SN
Sbjct 271 NPKDARLQRPEYMGGGTTQINFSEVLQTSPEIPGEDQVSQFG--VGDMYG-HGIAAMRSN 327
Query 179 NIEVFCDEESYIIGLLTVTPVPVYTQLLPKDFVYNGLLDHYQPEFDRIGFQPITYKEICP 238
+ +E YII +L+V P +YT + + ++ D+YQ E + IG Q I EI
Sbjct 328 KYRRYIEEHGYIISMLSVRPKTMYTNGIHRSWLRLTKEDYYQKELEHIGQQEIMNNEIYA 387
Query 239 MNIVGDDDTEQLSKTFGYQRPWYEYVAKYDSAHGLFRKDMKNFIMSRVFKGLPQLGQQFL 298
G ++TFGY + EY FR + + M+R F+ P L Q F
Sbjct 388 DEGAG-------TETFGYNDRYSEYRETPSHVSAEFRGILNYWHMAREFEAPPVLNQSF- 439
Query 299 LVDSDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 340
VD D ++ + + D ++ ++ AR +SR A PR+
Sbjct 440 -VDCDATKRIHN-EQTQDALWIMIQHKMVARRLLSRNAAPRI 479
Lambda K H a alpha
0.319 0.138 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1989760843275