bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_1
Length=523
Score E
Sequences producing significant alignments: (Bits) Value
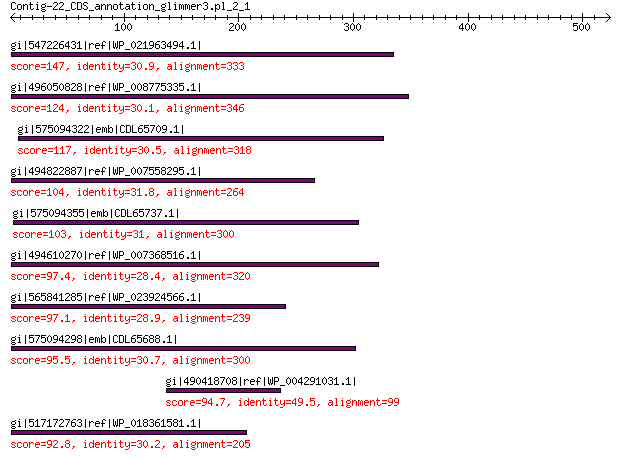
gi|547226431|ref|WP_021963494.1| predicted protein 147 4e-35
gi|496050828|ref|WP_008775335.1| hypothetical protein 124 2e-27
gi|575094322|emb|CDL65709.1| unnamed protein product 117 3e-25
gi|494822887|ref|WP_007558295.1| hypothetical protein 104 1e-20
gi|575094355|emb|CDL65737.1| unnamed protein product 103 4e-20
gi|494610270|ref|WP_007368516.1| hypothetical protein 97.4 3e-18
gi|565841285|ref|WP_023924566.1| hypothetical protein 97.1 4e-18
gi|575094298|emb|CDL65688.1| unnamed protein product 95.5 1e-17
gi|490418708|ref|WP_004291031.1| hypothetical protein 94.7 1e-17
gi|517172763|ref|WP_018361581.1| hypothetical protein 92.8 1e-16
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 147 bits (370), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 103/340 (30%), Positives = 163/340 (48%), Gaps = 44/340 (13%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
++NKYTG+ + V CG C+ C+ +A K C ++ + KYC F TLTYS +Y+P Y
Sbjct 17 VQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYSNDYVP--RMY 74
Query 62 KGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYT 121
E+R L R + Y + + + ++ + +Y+ L L K
Sbjct 75 PEVDNELR---LVRWYSYCDRLNEK-GKLMTVDYDYWHKCPSLDTYVLMLTAKC------ 124
Query 122 SFPDGRRIYNRPYMENLIGYLNY---RDMQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTT 178
NL GYL+Y RD QLF KR+ +N+ ++EKI YY+V EYGP T
Sbjct 125 ---------------NLDGYLSYTSKRDAQLFLKRVRKNLSKYSDEKIRYYIVSEYGPKT 169
Query 179 FRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNSTACLPDFY 238
FR H+H+L F+D + ++ + + + ++W+FG D + YVA YVN CLP F
Sbjct 170 FRAHYHVLFFYDEVKTQKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLPRFL 229
Query 239 KNFSHIKPFG----RFSMHFAESAFNEVFKPQEDEEIFSLFYDGRMLELNGKPTLVRPKR 294
+ S KPF RF++ +S E++K D+ I+ + E+NG P R
Sbjct 230 GDMS-TKPFSCHSIRFALGIHQSQKEEIYKGSVDDFIY------QSGEINGNYVEFMPWR 282
Query 295 SHINRLYPRLNKSKHASVDDDIRVATALSNIPHVLAKFGF 334
+ +P K K S D + + + + V + G+
Sbjct 283 NLSCTFFP---KCKGYSRKSDSELWQSYNILREVRSAIGY 319
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 124 bits (311), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 104/350 (30%), Positives = 154/350 (44%), Gaps = 50/350 (14%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
I N YT + + VPCG C+ C K + +C+++ +K+ FITLTY+ ++P F
Sbjct 15 IMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFITLTYANRFIPRAMFV 74
Query 62 KGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYT 121
S E + C + D +T A L +
Sbjct 75 D--SIERPYGC-----------------------DLIDKETGEILGPADLTED------- 102
Query 122 SFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNI-RSVTNEKIYYYVVGEYGPTTFR 180
+ + N+ Y+ + YL D+QLF KRL + + +EK+ Y+ VGEYGP FR
Sbjct 103 ---ERTNLLNKFYLFGDVPYLRKTDLQLFLKRLRYYVTKQKPSEKVRYFAVGEYGPVHFR 159
Query 181 PHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKN 240
PH+H+LLF S E Q + +SK+W FG D Q S YVA YVNS+ +P +K
Sbjct 160 PHYHLLLFLQSDEALQICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKA 219
Query 241 FSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGRM---LELNGKPTLVRPKRSHI 297
S + PF S + F + E+I+SL + + + LNGK RS
Sbjct 220 -SSVCPFSVHSQKLGQG-----FLDCQREKIYSLTPENFIRSSIVLNGKYKEFDVWRSCY 273
Query 298 NRLYPRLNKSKHASVDDDIRVATALSNIPHVLAKFGFIDEVTDFEMSKRI 347
+ YPR S + A S + A+ F D T F ++K I
Sbjct 274 SFFYPRCKGFVTKSSRE-----RAYSYSIYDTARLLFPDAKTTFSLAKEI 318
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 117 bits (294), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 97/341 (28%), Positives = 145/341 (43%), Gaps = 44/341 (13%)
Query 8 GDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFYKGASGE 67
G P VPCG C C +NK LK ++ SKYC F+TLTY + LP+
Sbjct 19 GYPYQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTYDDDNLPLFSVGLDTCAT 78
Query 68 VRFRCLPRDFVYSYKTVQGYNRNISFNDEYF--------DFDTQLSWESAQLLQKKTHLH 119
R P YS + RN SF ++ DF ++ + S ++ ++ H
Sbjct 79 EFVRIYP----YSERL-----RNDSFISDFCSDLHNFDNDFVDKMDYYSDYVINYESKYH 129
Query 120 YTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTF 179
+ Y L L YRD+QLF KRL ++I EKI +Y++GEYG +
Sbjct 130 KSCV----------YGHGLYALLYYRDIQLFLKRLRKHIYKYYGEKIRFYIIGEYGTKSL 179
Query 180 RPHFHILLFHDSKELRQSIR---------------QFVSKSWRFGDTDTQPVWSSASCYV 224
RPH+H LLF +S L Q+ +F+ W+FG D++ A YV
Sbjct 180 RPHWHCLLFFNSSSLSQAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGEAYNYV 239
Query 225 AGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGRMLELN 284
+ YVN +A P S+ K + + S + V Q+ + FS F L+
Sbjct 240 SSYVNQSANFPKLLVLLSNQKAYHSIQLGQILSEQSIVSAIQKGD--FSFFERQFYLDTF 297
Query 285 GKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNI 325
G RS+ +R +P+ S + + RV T +
Sbjct 298 GAANSYSVWRSYYSRFFPKFTCSSQLTYEQTYRVLTCYETL 338
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 104 bits (260), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 84/294 (29%), Positives = 117/294 (40%), Gaps = 64/294 (22%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
IKNKYTG+ + V C C C + K C+ + +K F+TLT+ +++P FY
Sbjct 14 IKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDDKFVPQFRFY 73
Query 62 KGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYT 121
K E R D DT + L+ + Y
Sbjct 74 KVGDDEYIMR---------------------------DADTG-EYLGRTLMTPQLMNEYQ 105
Query 122 SFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRP 181
+R+ R + YL+ R++QLF KRL + + +KI ++ GEYGP +FRP
Sbjct 106 -----KRVNYRINYKGRFPYLSKRELQLFMKRLRKYLDKYEGQKIRFFATGEYGPLSFRP 160
Query 182 HFHILLFHDSKE-----------------------------LRQSIRQFVSKSWRFGDTD 212
HFHILLF D L + ++ +SW FG D
Sbjct 161 HFHILLFVDDPSLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLSKLEYYIRESWPFGGID 220
Query 213 TQPV-WSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKP 265
Q V S S YVAGYVNS+ LP K +K F + S F P
Sbjct 221 AQSVEQGSCSSYVAGYVNSSVPLPSCLK-VDAVKSFSQHSRFLGRKIFGTELIP 273
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 103 bits (256), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 93/331 (28%), Positives = 140/331 (42%), Gaps = 68/331 (21%)
Query 4 NKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFYKG 63
N Y D + VPCG C C +KA + +L+ ++ + K+C F TLTY+ Y+P
Sbjct 19 NPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYANTYIP------- 71
Query 64 ASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYTSF 123
R +P + ++ V GY EY + S++ LL K HL F
Sbjct 72 -----RLSLVPYN-DKTFGVVNGYEMCDKETGEYLGYLDSPSYDVESLLDK-LHL----F 120
Query 124 PDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHF 183
D + YL RD+QLF KRL +N+ ++ K+ Y+ +GEYGP FRPH+
Sbjct 121 GD-------------VPYLRKRDLQLFIKRLRKNLSKYSDAKVRYFAMGEYGPVHFRPHY 167
Query 184 HILLFHDS----------------------------KELRQSIRQFVSKSWRFGDTDTQP 215
H LLF D ++ + + SW+FG D Q
Sbjct 168 HFLLFFDEIKFTAPSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWKFGRVDAQY 227
Query 216 VWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSL- 274
A+ YV+ YV+ + LP Y+ S +PF S + F E E+++
Sbjct 228 SKGDAAQYVSSYVSGSGSLPKVYQ-VSSARPFSLHSRFLGQG-----FLAHECEKVYETP 281
Query 275 --FYDGRMLELNGKPTLVRPKRSHINRLYPR 303
+ R +ELNG RS + YP+
Sbjct 282 VRDFVKRSVELNGSNKDFNLWRSCYSVFYPK 312
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 97.4 bits (241), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 91/337 (27%), Positives = 147/337 (44%), Gaps = 66/337 (20%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
I NKY + +YVPC C C + A + + ++ F+TLTY E++P+
Sbjct 16 IYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRFSLFVTLTYDNEHIPL---- 71
Query 62 KGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYT 121
F D + W S +L + L +
Sbjct 72 -------------------------------FQPLVMDDGSHPVWFSNRLSESGKFLSDS 100
Query 122 ---SFPDGRRIYNRPYMENLI--GYLNYRDMQLFFKRLNQNI-----RSVTNE-KIYYYV 170
S P + ME+ + Y +D+Q +FKRL + ++ +NE +I Y++
Sbjct 101 VCRSLPPQK-------MEDEVCFAYPCKKDVQDWFKRLRSAVDYQLNKNKSNEFRIRYFI 153
Query 171 VGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNS 230
EYGP TFRPH+H +L++DS+EL+++I + + ++W+ G++ V +SAS YVA YVN
Sbjct 154 CSEYGPRTFRPHYHAILWYDSEELQRNIGRLIRETWKNGNSVFSLVNNSASQYVAKYVNG 213
Query 231 TACLPDFYKN-FSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGR-----MLELN 284
LP F + F+ + H A + ++E + S DG + N
Sbjct 214 DTRLPPFLRTEFTS-------TFHLASKHPYIGYCKADEEALRSNVLDGTYGQSVLNRDN 266
Query 285 GKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATA 321
G+ V RS NRL P+ + S + IRV A
Sbjct 267 GQFEFVPTPRSLENRLLPKCRGYRSLSHSERIRVYAA 303
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 97.1 bits (240), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 69/246 (28%), Positives = 109/246 (44%), Gaps = 47/246 (19%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
I N YT + V+V C C+ C++ K + + Y F+TLTY E+LP+
Sbjct 15 IINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSAFVTLTYDNEHLPL---- 70
Query 62 KGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHLHYT 121
+ E + ++ W S +L +K +
Sbjct 71 -------------------------------YQPECMNERGEMVWTSNRLCDEKVIVGNY 99
Query 122 SFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNI-------RSVTNEKIYYYVVGEY 174
F ++ N + Y D+ FFKRL + +TNEKI Y+V EY
Sbjct 100 DFI---KVSNSDVQA--VAYCCKSDIVKFFKRLRSKLSYYFKKHHIITNEKIRYFVCSEY 154
Query 175 GPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNSTACL 234
GP T RPH+H +++ DS+E+ + I + +S SW G TD + V S+A YVA YV+ + L
Sbjct 155 GPKTLRPHYHAIIWFDSEEVARVIEKMLSSSWSNGFTDFEYVNSTAPQYVAKYVSGNSVL 214
Query 235 PDFYKN 240
P+ ++
Sbjct 215 PEILQH 220
>gi|575094298|emb|CDL65688.1| unnamed protein product [uncultured bacterium]
Length=478
Score = 95.5 bits (236), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 92/328 (28%), Positives = 139/328 (42%), Gaps = 46/328 (14%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPV---G 58
I+NKYTG +YV CG C C+ KA + K ++ C F+TL Y ++PV
Sbjct 8 IRNKYTGQKLYVSCGKCPACLQEKANASAYKIRNNQSSELSCFFVTLNYDNNHIPVIFKH 67
Query 59 EFYKGASGEV-------RFRCLPRDFVYSYKTVQGYNR----NISFNDEYFDFDTQLSWE 107
+ Y S +V + CLP D +Y N+ N N D + L
Sbjct 68 DVYNYNSSDVYHFDEERKELCLPVD-LYRGVCPAFSNKIDTFNFPLNRLSTDVVSSLDNH 126
Query 108 SAQLLQKKTHLHYTSFPDGRRIYNRPYM--ENLIGYLNYRDMQLFFKRLNQNIRSVTNEK 165
+++ K H +P + E + +D+QLFFKRL Q++ +
Sbjct 127 CGVVVKTKNH--------------KPVLFNEEIFSVCYTKDIQLFFKRLRQSLYRKFGFR 172
Query 166 --IYYYVVGEYGPTTFRPHFHILLFHDSKELR-QSIRQFVSKSWRFGDTDTQ----PVWS 218
I Y+ EYGPTT+R HFH+ +F E+ S R+ K+W F +
Sbjct 173 PFIQYFQTSEYGPTTYRAHFHLCIFVKRSEISFDSFRKACVKAWPFCSKKQMFRNVEIAR 232
Query 219 SASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAES----AFNEVFKPQEDEEIFSL 274
S S Y+A YVN A +P F N K S++F + +FN + E + +
Sbjct 233 SPSAYIASYVNCRANVPLFL-NLKEAKAKHTHSLYFGHNNDKLSFNSIVNRFETQG--TT 289
Query 275 FYDGRMLELNGKP-TLVRPKRSHINRLY 301
Y + ++G P T P +IN Y
Sbjct 290 LYPRELSSVDGVPQTSFLPLPRYINAYY 317
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 94.7 bits (234), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 49/103 (48%), Positives = 62/103 (60%), Gaps = 4/103 (4%)
Query 137 NLIGYLNYR---DMQLFFKRLNQNI-RSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSK 192
+L GYL Y D+QLFFKR + + EK+ Y+ +GEYGP FRPH+HILLF S
Sbjct 36 HLFGYLPYLRKFDLQLFFKRFRYYVAKRFPKEKVRYFAIGEYGPVHFRPHYHILLFLQSD 95
Query 193 ELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYVNSTACLP 235
E Q + VS++W FG D Q S YVAGYVNS+ +P
Sbjct 96 EALQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVP 138
>gi|517172763|ref|WP_018361581.1| hypothetical protein [Prevotella nanceiensis]
Length=598
Score = 92.8 bits (229), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 62/212 (29%), Positives = 100/212 (47%), Gaps = 31/212 (15%)
Query 2 IKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYLPVGEFY 61
I NK TG VPC C +C++ +A K + ++ + TLTY Y+P E +
Sbjct 32 IYNKNTGHYETVPCHNCTYCVNVEASKQSRRVREEIKQHLFSVMFTLTYDNVYIPRMEAF 91
Query 62 KGASGEVRFRCLPR--DFVYSYK-TVQGYNRNISFNDEYFDFDTQLSWESAQLLQKKTHL 118
G GE++ + + R D S + YN + FND DT++ W + K +L
Sbjct 92 AGKHGEMQLKPIGRTADLHDSCPFNSKNYNGDYRFND-----DTRIPWIENNKIYCKNNL 146
Query 119 HYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVT----NEKIYYYVVGEY 174
+ + ++ +D+Q F KRL + I + +KI Y++ EY
Sbjct 147 QFAT-------------------VSKKDIQNFLKRLRKKIDKLNIPQNEKKIRYFIASEY 187
Query 175 GPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 206
GP T+RPH+H +LF DS + I+ F+ +SW
Sbjct 188 GPKTYRPHYHGVLFIDSPTVLSKIKAFIVESW 219
Lambda K H a alpha
0.323 0.138 0.424 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3723896675700