bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_4
Length=193
Score E
Sequences producing significant alignments: (Bits) Value
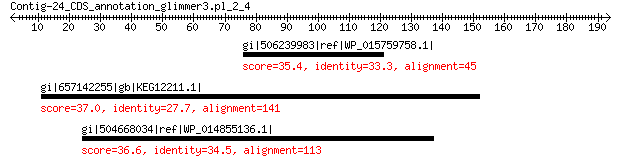
gi|506239983|ref|WP_015759758.1| hypothetical protein 35.4 6.0
gi|657142255|gb|KEG12211.1| putative phosphatidylinositol 3-kinase 37.0 7.8
gi|504668034|ref|WP_014855136.1| hypothetical protein 36.6 8.7
>gi|506239983|ref|WP_015759758.1| hypothetical protein [Alicyclobacillus acidocaldarius]
gi|257793022|ref|YP_003186421.1| hypothetical protein Aaci_3029 [Alicyclobacillus acidocaldarius
subsp. acidocaldarius DSM 446]
gi|257479714|gb|ACV60032.1| hypothetical protein Aaci_3029 [Alicyclobacillus acidocaldarius
subsp. acidocaldarius DSM 446]
Length=98
Score = 35.4 bits (80), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 76 SKKIAAFVLLHDSLTALDFSSFHLPLKKVGKETFLSSLMESACRS 120
++K A +L HD LT L+++SFH+ +K+ L++ + A R+
Sbjct 11 ARKRAEELLTHDDLTMLEYNSFHIIVKQSDTNEILANALREAVRA 55
>gi|657142255|gb|KEG12211.1| putative phosphatidylinositol 3-kinase [Trypanosoma grayi]
Length=2814
Score = 37.0 bits (84), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 39/141 (28%), Positives = 64/141 (45%), Gaps = 8/141 (6%)
Query 11 STDPSLSAHISISYFSLTPTNSPRILSELLIHVGNLDVSIALRPEERLSRTLRDILIVLH 70
S D L I + L+ +++P++L + + L V P+++ +R R + IVLH
Sbjct 133 SRDLLLEQSIRDALSELSRSSNPKLLLLAVDFIDALLVVPYTNPQQKFTRLCRSLFIVLH 192
Query 71 LFPYISKKIAAFVLLHDSLTALDFSSFHLPLKKVGKETFLSSLMESACRSMASIELFGHG 130
K A VL L FS F PLK +F++ ++ C +A I+L G
Sbjct 193 CGIEKPAKEAKRVLKRMLAMDLHFSPFDPPLK-----SFITKELKDNC-ELALIQLHGRA 246
Query 131 ARLSIPAFFDPSHIAVVLYLN 151
+ P F +AV+L L+
Sbjct 247 QQTKAP--FVAQFLAVMLALD 265
>gi|504668034|ref|WP_014855136.1| hypothetical protein [Melioribacter roseus]
gi|397689458|ref|YP_006526712.1| hypothetical protein MROS_0456 [Melioribacter roseus P3M-2]
gi|395810950|gb|AFN73699.1| hypothetical protein MROS_0456 [Melioribacter roseus P3M-2]
Length=517
Score = 36.6 bits (83), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 39/140 (28%), Positives = 58/140 (41%), Gaps = 33/140 (24%)
Query 24 YFSLTPTNSPR-------ILSELLIHVGNLDVSIALRP---------EERLSRTLRDILI 67
YFSL R +S+LL+H + +V+ RP +E RT +I
Sbjct 73 YFSLINEEIKRNKKLSSDFVSKLLLHASSFNVNFLARPRWTLTKFIFDENNHRTGIEIKQ 132
Query 68 VLH-----------LFPYISKKIAAFVLLHDSLTALDFSSFHLPLKKVGKETFLSSLMES 116
+L+ +F YI KK SL + +FS + +GKET L SL+
Sbjct 133 ILNYLYYYSYLKKIIFSYIDKKKII------SLNSEEFSELLQKIDDIGKETNLQSLLSG 186
Query 117 ACRSMASIELFGHGARLSIP 136
A +SMA G + +P
Sbjct 187 ALKSMADFFNIGELQKNKVP 206
Lambda K H a alpha
0.323 0.137 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 580540099968