bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_4
Length=347
Score E
Sequences producing significant alignments: (Bits) Value
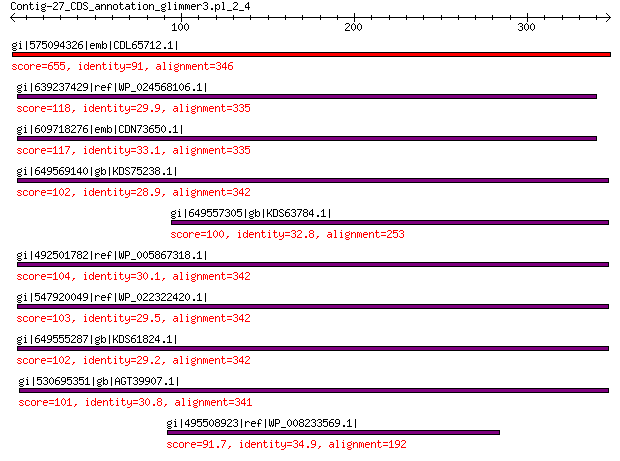
gi|575094326|emb|CDL65712.1| unnamed protein product 655 0.0
gi|639237429|ref|WP_024568106.1| hypothetical protein 118 3e-26
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 117 6e-26
gi|649569140|gb|KDS75238.1| capsid family protein 102 2e-21
gi|649557305|gb|KDS63784.1| capsid family protein 100 2e-21
gi|492501782|ref|WP_005867318.1| hypothetical protein 104 2e-21
gi|547920049|ref|WP_022322420.1| capsid protein VP1 103 3e-21
gi|649555287|gb|KDS61824.1| capsid family protein 102 9e-21
gi|530695351|gb|AGT39907.1| major capsid protein 101 2e-20
gi|495508923|ref|WP_008233569.1| capsid protein 91.7 3e-18
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 655 bits (1690), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 315/346 (91%), Positives = 326/346 (94%), Gaps = 0/346 (0%)
Query 2 YANWQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDF 61
+ANWQSDAYTTAL APQQ V PLVGLTTYE SVN+AGH VTTVNTAIVDEEGNAYKVDF
Sbjct 413 FANWQSDAYTTALTAPQQGVAPLVGLTTYEIRSVNDAGHEVTTVNTAIVDEEGNAYKVDF 472
Query 62 ESNGDALKGVNYTPLKAGEAVNMQSLVSPVASGISINDFRNVNAYQRYLELNQFRGFSYK 121
ESNG+ALKGVNYTPLKAGEAVNMQSLVSPV SGISINDFRNVNAYQRYLELNQFRGFSYK
Sbjct 473 ESNGEALKGVNYTPLKAGEAVNMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYK 532
Query 122 EIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQAGLATCF 181
EIIEGRFDVNVRYDALNMPEYLGGITRDI+VNPITQTVETT SGSYVGSLGSQ+GLATCF
Sbjct 533 EIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETTGSGSYVGSLGSQSGLATCF 592
Query 182 GNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIYL 241
GN+DGSISVFCDEESIVMGIMYVMPMPVYDS+LPKWLTYRERLDSFNPEFDHIGYQPIY
Sbjct 593 GNTDGSISVFCDEESIVMGIMYVMPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYA 652
Query 242 KELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELGK 301
KEL +Q + D NTVFGYQRPWYEYV K DRAHGLFLSSLRNFIMFRSF+N PELG+
Sbjct 653 KELGPMQCVQDDIDPNTVFGYQRPWYEYVAKPDRAHGLFLSSLRNFIMFRSFDNVPELGQ 712
Query 302 SFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 347
SFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE
Sbjct 713 SFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 758
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 118 bits (296), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 100/352 (28%), Positives = 162/352 (46%), Gaps = 33/352 (9%)
Query 5 WQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTV------TTVNTAIVDEEGNA-- 56
W D +T+AL Q+ + L + N G+T + +T E+G+
Sbjct 202 WHHDYFTSALPFAQKGAAVKMPLQMTADLFYNPGGNTFVKKPDGSLSHTGFRLEDGSVPA 261
Query 57 -----YKVDFESNGDALKGVNYTPLKAGEAVNMQSLVSPVASGISINDFRNVNAYQRYLE 111
V+ S G N P+ + N+ + ASG +IND R Q +LE
Sbjct 262 DGIGHLMVETSSTG------NSNPVNIDNSSNLGVDLK-TASGSTINDLRRAFKLQEWLE 314
Query 112 LNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSL 171
N G Y E I F V L PE+LGG I+++ + Q +TDS + G++
Sbjct 315 KNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILISEVLQQ-SSTDSTTPQGNM 373
Query 172 GSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEF 231
AG G +G S F +E V+G+M V+P Y +P+ + ++ D F P+F
Sbjct 374 ---AGHGISVGK-EGGFSKFFEEHGYVIGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQF 429
Query 232 DHIGYQPIYLKELAA--IQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIM 289
+HIG QP+Y KE+ A + ++SG VFGY + EY HG F +L + +
Sbjct 430 EHIGEQPVYNKEIFAKNVGDYDSG----GVFGYVPRYSEYKYSPSTIHGDFKDTLYFWHL 485
Query 290 FRSFENA--PELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPIS 339
R F+++ P+L + F + ++ +F+V + SDK ++ TA+ +S
Sbjct 486 GRIFDSSAPPKLNRDFIEVNKSGLSRIFAVEDNSDKFYCHLYQKITAKRKMS 537
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 117 bits (293), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 111/354 (31%), Positives = 162/354 (46%), Gaps = 46/354 (13%)
Query 5 WQSDAYTTALAAPQ-----------QDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEE 53
W D +T+AL Q Q VPL TYE S T I D
Sbjct 202 WHHDYFTSALPFAQKGNAVKIPIFPQGNVPL----TYEMGS-----------QTFIKDMA 246
Query 54 GN-AYKVDFES--NGDALKGVNYTPLKAGEAVNMQ-SLVSPVASGISINDFRNVNAYQRY 109
GN A D S NG+ L+ V+ PL + N++ ++ S S ++ND R Q +
Sbjct 247 GNPAPNKDLRSDVNGN-LQDVSGQPLSLDPSKNLKLNMASENVS--TVNDLRRAFKLQEW 303
Query 110 LELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVG 169
LE N G Y E I F V L PE+LGG I+++ + Q TDS + G
Sbjct 304 LEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMISEVLQQ-SATDSTTPQG 362
Query 170 SLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNP 229
++ AG G DG S F +E V+G+M V+P Y +P+ + ++ D F P
Sbjct 363 NM---AGHGIGIG-KDGGFSRFFEEHGYVIGLMSVIPKTSYSQGIPRHFSKSDKFDYFWP 418
Query 230 EFDHIGYQPIYLKELAA--IQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNF 287
+F+HIG QP+Y KE+ A I AF+S VFGY + EY HG F L +
Sbjct 419 QFEHIGEQPVYNKEIFAKNIDAFDS----EAVFGYLPRYSEYKFSPSTVHGDFKDDLYFW 474
Query 288 IMFRSF--ENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPIS 339
+ R F + P L +SF +++ +F+V + +DK ++ TA+ +S
Sbjct 475 HLGRIFDTDKPPVLNQSFIECDKNALSRIFAVEDDTDKFYCHLYQKITAKRKMS 528
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 102 bits (255), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 99/352 (28%), Positives = 149/352 (42%), Gaps = 28/352 (8%)
Query 5 WQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDFESN 64
W+ D +T+AL P V TV +N G + ++ + +
Sbjct 57 WEKDYFTSAL--------PWVQRGPEVTVPINGGGEIPVEMKEGFAAQKITTFPDRKPIS 108
Query 65 GDALKGVNYTPLKAGE--AVNMQSLVSPV-------ASGISINDFRNVNAYQRYLELNQF 115
G + + L G+ ++ Q+L+ P G++IND R NA QR+ E N
Sbjct 109 GSEVLYSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDIRTSNALQRWFERNAR 168
Query 116 RGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQA 175
G Y E I F V L P++LGG I V+ + QT +TDS S ++ A
Sbjct 169 SGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQT-SSTDSTSPQANM---A 224
Query 176 GLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIG 235
G G + G + + +E +MGIM + P Y +PK + +D + PEF H+G
Sbjct 225 GHGISAGVNHG-FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLG 283
Query 236 YQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFEN 295
Q I +EL ES FGY + EY + HG F ++ + + R F+
Sbjct 284 EQEIKNEELYLN---ESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKE 340
Query 296 APELGKSFTVMQPGSVNNVFSVTEVS-DKILGQIHFDCTAQLPISRVVVPRL 346
P L +F P N VF+ E S DK QI+ D A + + P L
Sbjct 341 KPNLNTTFVECNPS--NRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 83/254 (33%), Positives = 115/254 (45%), Gaps = 11/254 (4%)
Query 94 GISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVN 153
G++IND R NA QR+ E N G Y E I F V L P++LGG I V+
Sbjct 2 GVNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVS 61
Query 154 PITQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSI 213
+ QT +TDS S ++ AG G + G F +E +MGIM + P Y
Sbjct 62 EVLQT-SSTDSTSPQANM---AGHGISAGVNHGFTRYF-EEHGYIMGIMSIRPRTGYQQG 116
Query 214 LPKWLTYRERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKV 273
+PK + +D + PEF H+G Q I +EL ES FGY + EY
Sbjct 117 VPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLN---ESDAANEGTFGYTPRYAEYKYSQ 173
Query 274 DRAHGLFLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVS-DKILGQIHFDC 332
+ HG F ++ + + R F+ P L +F P N VF+ E S DK QI+ D
Sbjct 174 NEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS--NRVFATAETSDDKYWVQIYQDI 231
Query 333 TAQLPISRVVVPRL 346
A + + P L
Sbjct 232 KALRLMPKYGTPML 245
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 104 bits (259), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 103/351 (29%), Positives = 151/351 (43%), Gaps = 31/351 (9%)
Query 5 WQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDFESN 64
W+ D +T+AL P V TV + +G + T D + Y++ SN
Sbjct 210 WEKDYFTSAL--------PWVQRGPEVTVPIQGSGGNLDV--TLKNDAHADTYRMPGTSN 259
Query 65 --GDALKGVNYTPLKAG------EAVNMQSLVSPVASGISINDFRNVNAYQRYLELNQFR 116
A++ V + G E N Q V + G+SIND R NA QR+ E N
Sbjct 260 RPAGAMQLVGGALIAGGTDGAYLEPDNFQVNVDEL--GVSINDLRTSNALQRWFERNARS 317
Query 117 GFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQAG 176
G Y E I F V L P++LGG I V+ + QT TDS S ++ AG
Sbjct 318 GSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQT-SATDSTSPQANM---AG 373
Query 177 LATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGY 236
G + G + +E ++GIM + P Y +PK + +D + PEF H+G
Sbjct 374 HGISAGVNHG-FKRYFEEHGYIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGE 432
Query 237 QPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENA 296
Q I +E+ Q S N FGY + EY ++ HG F ++ + + R F +
Sbjct 433 QEIKNEEVYLQQTPASN---NGTFGYTPRYAEYKYSMNEVHGDFRGNMAFWHLNRIFSES 489
Query 297 PELGKSFTVMQPGSVNNVFSVTEVS-DKILGQIHFDCTAQLPISRVVVPRL 346
P L +F P N VF+ E S DK Q++ D A + + P L
Sbjct 490 PNLNTTFVECNPS--NRVFATAETSDDKYWIQLYQDVKALRLMPKYGTPML 538
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 103 bits (258), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 101/361 (28%), Positives = 152/361 (42%), Gaps = 37/361 (10%)
Query 5 WQSDAYTTALAAPQQD---VVPLVGLTTYETVSVNEAGHTVTTVNTAIVD-EEGNAYKVD 60
W+ D +T+AL Q+ VP+ G V + V+++ + E G+AY +
Sbjct 211 WEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSDSQKWVDSSGREFENGHAYDIT 270
Query 61 F----ESNGDALKGVNYTPLKAGEAVNMQSLVSPVAS--------GISINDFRNVNAYQR 108
+ N + VN G N + P + GI+IND R NA QR
Sbjct 271 MARANDPNSALMVAVN------GGTNNRAPELDPNGTLKVNVDEMGININDLRTSNALQR 324
Query 109 YLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVET--TDSGS 166
+ E N G Y E I F V L P++LGG I V+ + QT T T +
Sbjct 325 WFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQA 384
Query 167 YVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDS 226
+ G AG+ F + + +E ++GIM + P Y +P+ T + +D
Sbjct 385 NMAGHGISAGINNGFKH-------YFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDF 437
Query 227 FNPEFDHIGYQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRN 286
+ PEF H+ Q I +EL E N FGY + EY AHG F +L
Sbjct 438 YFPEFAHLSEQEIKNQELFVS---EDAAYNNGTFGYTPRYAEYKYHPSEAHGDFRGNLSF 494
Query 287 FIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVS-DKILGQIHFDCTAQLPISRVVVPR 345
+ + R FE+ P L +F +P N VF+ +E DK Q++ D A + + P
Sbjct 495 WHLNRIFEDKPNLNTTFVECKPS--NRVFATSETEDDKFWVQMYQDVKALRLMPKYGTPM 552
Query 346 L 346
L
Sbjct 553 L 553
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 102 bits (254), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 100/352 (28%), Positives = 148/352 (42%), Gaps = 28/352 (8%)
Query 5 WQSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHTVTTVNTAIVDEEGNAYKVDFESN 64
W+ D +T+AL P V TV +N G + ++ + +
Sbjct 208 WEKDYFTSAL--------PWVQRGPEVTVPINGGGEIPVEMKEGFAAQKITTFPDRKPIS 259
Query 65 GDALKGVNYTPLKAGE--AVNMQSLVSPV-------ASGISINDFRNVNAYQRYLELNQF 115
G + + L G+ ++ Q+L+ P G++IND R NA QR+ E N
Sbjct 260 GSEVLYSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDIRTSNALQRWFERNAR 319
Query 116 RGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQA 175
G Y E I F V L P++LGG I V+ + QT +TDS S ++ A
Sbjct 320 SGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQT-SSTDSTSPQANM---A 375
Query 176 GLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIG 235
G G + G F +E +MGIM + P Y +PK + +D + PEF H+G
Sbjct 376 GHGISAGVNHGFTRYF-EEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLG 434
Query 236 YQPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFEN 295
Q I +EL ES FGY + EY + HG F ++ + + R F+
Sbjct 435 EQEIKNEELYLN---ESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKE 491
Query 296 APELGKSFTVMQPGSVNNVFSVTEVS-DKILGQIHFDCTAQLPISRVVVPRL 346
P L +F P N VF+ E S DK QI+ D A + + P L
Sbjct 492 KPNLNTTFVECNPS--NRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 541
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 101 bits (252), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 105/356 (29%), Positives = 154/356 (43%), Gaps = 30/356 (8%)
Query 6 QSDAYTTALAAPQQDVVPLVGLTTYETVSVNEAGHT--VTTVNTAIVDEEGNAYKVDFES 63
+ D +T+AL PQ+ + L V N+ G + V+T V + V E+
Sbjct 195 RHDYFTSALPWPQKGDAVTLPLGGSANVVYNDTGDPAYIREVSTGNVWTTPSRESVSKEA 254
Query 64 NGDALKGVNYTPLKAGEAVNMQ-----SLVSPV--ASGISINDFRNVNAYQRYLELNQFR 116
NG+ + G +VN Q SLV+ + A+ +IN R QR LE +
Sbjct 255 NGN-------MSVPTG-SVNAQYDPNGSLVADLSTATAATINAIRQSFQIQRLLERDARG 306
Query 117 GFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIIVNPITQTVETTDSGSYVGSLGSQAG 176
G Y EI+ F V + PEYLGG + IIVNP+ Q + SG+ LG+
Sbjct 307 GTRYTEIVRSHFGVISPDARMQRPEYLGGGSAPIIVNPVAQQSASGASGTDT-PLGTLGA 365
Query 177 LATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGY 236
+ T + G S F E +V+G+ V Y L + + R D F P F H+G
Sbjct 366 VGTGLASGHGFASSFT-EHGVVVGLCSVRADLTYQQGLHRMFSRSTRYDFFFPVFSHLGE 424
Query 237 QPIYLKELAAIQAFESGKDLNTVFGYQRPWYEYVQKVDRAHGLFLS----SLRNFIMFRS 292
QPI KEL A + VFGYQ W EY K + GL S +L + + ++
Sbjct 425 QPILNKELYATGTSTD----DDVFGYQEAWAEYRYKPSQVTGLMRSTAAGTLDAWHLAQN 480
Query 293 FENAPELGKSFTVMQPGSVNNVFSVTEVSD--KILGQIHFDCTAQLPISRVVVPRL 346
F + P L +F P V+ V +V ++ + + FD P+ VP L
Sbjct 481 FGSLPTLNSTFIEDTP-PVDRVVAVGSEANGQQFIFDAFFDINMARPMPMYSVPGL 535
>gi|495508923|ref|WP_008233569.1| capsid protein [Richelia intracellularis]
gi|471330407|emb|CCH67259.1| capsid protein [Richelia intracellularis HH01]
Length=230
Score = 91.7 bits (226), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 67/193 (35%), Positives = 91/193 (47%), Gaps = 8/193 (4%)
Query 92 ASGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDII 151
A+ +IN RN QR LE + G Y EI+ F V L PEYLGG + I
Sbjct 15 ATSATINAIRNSFQIQRLLERDARGGTRYTEIVRSHFGVISPDARLQRPEYLGGGSAPIT 74
Query 152 VNPITQTVETTDSGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYD 211
VNPI QT +T +GS LG+ + T N G + F E +++G+ V Y
Sbjct 75 VNPIAQTSASTVTGSDT-PLGALGAVGTGLANGHGFSTSFT-EHGVILGLASVRADLTYQ 132
Query 212 SILPKWLTYRERLDSFNPEFDHIGYQPIYLKEL-AAIQAFESGKDLNTVFGYQRPWYEYV 270
L + + + R D + P F H+G Q + KE+ A +SG VFGYQ W EY
Sbjct 133 QGLHRMWSRQTRYDFYFPVFAHLGEQAVLNKEIYCDGTANDSG-----VFGYQERWAEYR 187
Query 271 QKVDRAHGLFLSS 283
K + GL S+
Sbjct 188 YKPSQVTGLMRST 200
Lambda K H a alpha
0.318 0.135 0.396 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2051618693325