bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-28_CDS_annotation_glimmer3.pl_2_2
Length=536
Score E
Sequences producing significant alignments: (Bits) Value
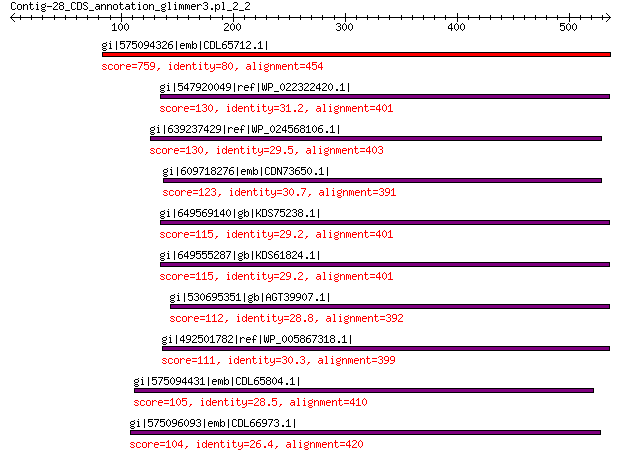
gi|575094326|emb|CDL65712.1| unnamed protein product 759 0.0
gi|547920049|ref|WP_022322420.1| capsid protein VP1 130 2e-29
gi|639237429|ref|WP_024568106.1| hypothetical protein 130 3e-29
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 123 6e-27
gi|649569140|gb|KDS75238.1| capsid family protein 115 7e-25
gi|649555287|gb|KDS61824.1| capsid family protein 115 4e-24
gi|530695351|gb|AGT39907.1| major capsid protein 112 3e-23
gi|492501782|ref|WP_005867318.1| hypothetical protein 111 1e-22
gi|575094431|emb|CDL65804.1| unnamed protein product 105 1e-20
gi|575096093|emb|CDL66973.1| unnamed protein product 104 2e-20
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 759 bits (1960), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 363/454 (80%), Positives = 408/454 (90%), Gaps = 2/454 (0%)
Query 83 LYLAVRQASQFWNSESNDHTLRFNYVGFDDFSVTNYYAGVTPYYGVGSTDHKAIKISAYP 142
+ L++ ++ N +S ++T + + FD V +G PYYG + KAIK+SAYP
Sbjct 307 ICLSLPSCTELCN-DSGNNTFKVG-ITFDKLDVFIGSSGKYPYYGSANMSDKAIKLSAYP 364
Query 143 FRAYEAVYNAYIRNTRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYANWQSDAYTSA 202
FRAYEA+YNAYIRNTRNNPF+LNGKKTYNR+IT DAGG+D+LTP+ L +ANWQSDAYT+A
Sbjct 365 FRAYEAIYNAYIRNTRNNPFVLNGKKTYNRWITTDAGGSDTLTPRDLRFANWQSDAYTTA 424
Query 203 LTAPQQGIAPLVGLTTYETKSINEAGHEVTVVNTAIVDEDGNAFKVDFESNGEALKGVNY 262
LTAPQQG+APLVGLTTYE +S+N+AGHEVT VNTAIVDE+GNA+KVDFESNGEALKGVNY
Sbjct 425 LTAPQQGVAPLVGLTTYEIRSVNDAGHEVTTVNTAIVDEEGNAYKVDFESNGEALKGVNY 484
Query 263 TPLKAGEALNMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVR 322
TPLKAGEA+NMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVR
Sbjct 485 TPLKAGEAVNMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVR 544
Query 323 YDALNMPEYLGGITRDIVVNPITQTVETSNTGAYVGALGSQAGLATCFGNTDGSISVFCD 382
YDALNMPEYLGGITRDIVVNPITQTVET+ +G+YVG+LGSQ+GLATCFGNTDGSISVFCD
Sbjct 545 YDALNMPEYLGGITRDIVVNPITQTVETTGSGSYVGSLGSQSGLATCFGNTDGSISVFCD 604
Query 383 EESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYVRELAAVQARQFN 442
EESIV+G+M+V+PMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIY +EL +Q Q +
Sbjct 605 EESIVMGIMYVMPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYAKELGPMQCVQDD 664
Query 443 VDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFIMYRSFVDVPELGKDFTTMKPGDVNN 502
+DP TVFGYQRPWYEYV KPDRAHGLFLSSLRNFIM+RSF +VPELG+ FT M+PG VNN
Sbjct 665 IDPNTVFGYQRPWYEYVAKPDRAHGLFLSSLRNFIMFRSFDNVPELGQSFTVMQPGSVNN 724
Query 503 VFSVTEVGDKILGQIHFDCTAQLPISRVVVPRLE 536
VFSVTEV DKILGQIHFDCTAQLPISRVVVPRLE
Sbjct 725 VFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 758
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 130 bits (328), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 125/426 (29%), Positives = 179/426 (42%), Gaps = 54/426 (13%)
Query 135 AIKISAYPFRAYEAVYNAYIRN---TRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFY 191
++SA PFRAY+ +YN Y R+ T F L T GG + SL
Sbjct 157 GFQVSALPFRAYQLIYNEYYRDQNLTEPIDFTLGSGTT--------VGGDQLMALMSLRR 208
Query 192 ANWQSDAYTSALTAPQQG---IAPLVGL-----TTYETKS-----INEAGHEVTVVNTAI 238
W+ D +TSAL Q+G P+ G YE +S ++ +G E
Sbjct 209 RAWEKDYFTSALPWLQRGPEVTVPVQGAGGSMDVVYERQSDSQKWVDSSGREF------- 261
Query 239 VDEDGNAFKVDF----ESNGEALKGVNYTPLKAGEAL--NMQSLVSPVTSGISINDFRNV 292
E+G+A+ + + N + VN L N V+ GI+IND R
Sbjct 262 --ENGHAYDITMARANDPNSALMVAVNGGTNNRAPELDPNGTLKVNVDEMGININDLRTS 319
Query 293 NAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSN 352
NA QR+ E N G Y E I F V L P++LGG I V+ + QT T
Sbjct 320 NALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDE 379
Query 353 TG--AYVGALGSQAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYR 410
T A + G AG+ F + + +E +IG+M + P Y +P+ T
Sbjct 380 TSPQANMAGHGISAGINNGFKH-------YFEEHGYIIGIMSITPRSGYQQGVPRDFTKF 432
Query 411 ERLDSFNPEFDHIGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFL 470
+ +D + PEF H+ Q I +EL + +N FGY + EY P AHG F
Sbjct 433 DNMDFYFPEFAHLSEQEIKNQELFVSEDAAYN---NGTFGYTPRYAEYKYHPSEAHGDFR 489
Query 471 SSLRNFIMYRSFVDVPELGKDFTTMKPGDVNNVFSVTEV-GDKILGQIHFDCTAQLPISR 529
+L + + R F D P L F KP N VF+ +E DK Q++ D A + +
Sbjct 490 GNLSFWHLNRIFEDKPNLNTTFVECKPS--NRVFATSETEDDKFWVQMYQDVKALRLMPK 547
Query 530 VVVPRL 535
P L
Sbjct 548 YGTPML 553
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 130 bits (326), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 119/429 (28%), Positives = 185/429 (43%), Gaps = 44/429 (10%)
Query 126 YGVGSTDHKAIKISAYPFRAYEAVYNAYIR--NTRNNPFILNGKKTYNRFITNDAGGADS 183
+ VG+ ++S PF AY+ +++ Y R N ++ F+ FI G +
Sbjct 127 FAVGNDGVLPDRVSMLPFLAYQKIWDEYYRDENLIDSVFVDKNGDKRELFID----GINY 182
Query 184 LTP------KSLFYAN---WQSDAYTSALTAPQQGIAPLVGLTTYETKSINEAGH----- 229
P + LF W D +TSAL Q+G A + L N G+
Sbjct 183 WNPSLPYEFRQLFDIKKRAWHHDYFTSALPFAQKGAAVKMPLQMTADLFYNPGGNTFVKK 242
Query 230 -EVTVVNTAIVDEDGNA-------FKVDFESNGEALKGVNYTPLKAGEALNMQSLVSPVT 281
+ ++ +T EDG+ V+ S G N P+ + N+ +
Sbjct 243 PDGSLSHTGFRLEDGSVPADGIGHLMVETSSTG------NSNPVNIDNSSNLGVDLK-TA 295
Query 282 SGISINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVV 341
SG +IND R Q +LE N G Y E I F V L PE+LGG I++
Sbjct 296 SGSTINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILI 355
Query 342 NPITQTVETSNTGAYVGALGSQAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDS 401
+ + Q T +T G+ AG G +G S F +E VIG+M VIP Y
Sbjct 356 SEVLQQSSTDSTTPQ----GNMAGHGISVGK-EGGFSKFFEEHGYVIGLMSVIPKTSYSQ 410
Query 402 LLPKWLTYRERLDSFNPEFDHIGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQK 461
+P+ + ++ D F P+F+HIG QP+Y +E+ A + D VFGY + EY
Sbjct 411 GIPRHFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVGDY--DSGGVFGYVPRYSEYKYS 468
Query 462 PDRAHGLFLSSLRNFIMYRSFVDV--PELGKDFTTMKPGDVNNVFSVTEVGDKILGQIHF 519
P HG F +L + + R F P+L +DF + ++ +F+V + DK ++
Sbjct 469 PSTIHGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNKSGLSRIFAVEDNSDKFYCHLYQ 528
Query 520 DCTAQLPIS 528
TA+ +S
Sbjct 529 KITAKRKMS 537
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 123 bits (309), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 120/411 (29%), Positives = 177/411 (43%), Gaps = 41/411 (10%)
Query 138 ISAYPFRAYEAVYNAYIRNTRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYA----- 192
++ PF AY+ +++ + R+ + + ND +L P S F
Sbjct 139 VNLLPFLAYQKIWDEFYRDENLIQPLFRDSNGNPVKMFNDGINDHNLPPYSKFTELFKMR 198
Query 193 --NWQSDAYTSALTAPQQGIAPLVGL-------TTYETKSINEAGHEVTVVNTAIVDEDG 243
W D +TSAL Q+G A + + TYE S T I D G
Sbjct 199 KRAWHHDYFTSALPFAQKGNAVKIPIFPQGNVPLTYEMGS-----------QTFIKDMAG 247
Query 244 N-AFKVDFES--NGEALKGVNYTPLKAGEALNMQ-SLVSPVTSGISINDFRNVNAYQRYL 299
N A D S NG L+ V+ PL + N++ ++ S S ++ND R Q +L
Sbjct 248 NPAPNKDLRSDVNGN-LQDVSGQPLSLDPSKNLKLNMASENVS--TVNDLRRAFKLQEWL 304
Query 300 ELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTGAYVGA 359
E N G Y E I F V L PE+LGG I+++ + Q T +T
Sbjct 305 EKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMISEVLQQSATDSTTPQ--- 361
Query 360 LGSQAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPE 419
G+ AG G DG S F +E VIG+M VIP Y +P+ + ++ D F P+
Sbjct 362 -GNMAGHGIGIGK-DGGFSRFFEEHGYVIGLMSVIPKTSYSQGIPRHFSKSDKFDYFWPQ 419
Query 420 FDHIGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFIMY 479
F+HIG QP+Y +E+ A F D VFGY + EY P HG F L + +
Sbjct 420 FEHIGEQPVYNKEIFAKNIDAF--DSEAVFGYLPRYSEYKFSPSTVHGDFKDDLYFWHLG 477
Query 480 RSF-VDVPE-LGKDFTTMKPGDVNNVFSVTEVGDKILGQIHFDCTAQLPIS 528
R F D P L + F ++ +F+V + DK ++ TA+ +S
Sbjct 478 RIFDTDKPPVLNQSFIECDKNALSRIFAVEDDTDKFYCHLYQKITAKRKMS 528
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 115 bits (288), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 117/416 (28%), Positives = 171/416 (41%), Gaps = 44/416 (11%)
Query 135 AIKISAYPFRAYEAVYNAYIRNTRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYAN- 193
K+SA PFRAY +YN Y R+ T IT D+G SL+ +
Sbjct 4 GFKVSALPFRAYHLIYNEYYRDQ---------NLTSELEITLDSGNYQLPVNSSLWQLHR 54
Query 194 --WQSDAYTSALTAPQQGIAPLVGLTTYETKSINEAGHEVTVVNTAIVDEDGNAFKVDFE 251
W+ D +TSAL Q+G P V T IN G + + F
Sbjct 55 RAWEKDYFTSALPWVQRG--PEV------TVPINGGGEIPVEMKEGFAAQKITTFPDRKP 106
Query 252 SNGEALKGVNYTPLKAGE--ALNMQSLVSPVT-------SGISINDFRNVNAYQRYLELN 302
+G + + L G+ ++ Q+L+ P G++IND R NA QR+ E N
Sbjct 107 ISGSEVLYSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDIRTSNALQRWFERN 166
Query 303 QFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTG--AYVGAL 360
G Y E I F V L P++LGG I V+ + QT T +T A +
Sbjct 167 ARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGH 226
Query 361 GSQAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEF 420
G AG+ F + + +E ++G+M + P Y +PK + +D + PEF
Sbjct 227 GISAGVNHGF-------TRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEF 279
Query 421 DHIGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFIMYR 480
H+G Q I EL ++ N FGY + EY + HG F ++ + + R
Sbjct 280 AHLGEQEIKNEELYLNESDAANEG---TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNR 336
Query 481 SFVDVPELGKDFTTMKPGDVNNVFSVTEVG-DKILGQIHFDCTAQLPISRVVVPRL 535
F + P L F P N VF+ E DK QI+ D A + + P L
Sbjct 337 IFKEKPNLNTTFVECNPS--NRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 115 bits (287), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 117/416 (28%), Positives = 171/416 (41%), Gaps = 44/416 (11%)
Query 135 AIKISAYPFRAYEAVYNAYIRNTRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYAN- 193
K+SA PFRAY +YN Y R+ T IT D+G SL+ +
Sbjct 155 GFKVSALPFRAYHLIYNEYYRDQ---------NLTSELEITLDSGNYQLPVNSSLWQLHR 205
Query 194 --WQSDAYTSALTAPQQGIAPLVGLTTYETKSINEAGHEVTVVNTAIVDEDGNAFKVDFE 251
W+ D +TSAL Q+G P V T IN G + + F
Sbjct 206 RAWEKDYFTSALPWVQRG--PEV------TVPINGGGEIPVEMKEGFAAQKITTFPDRKP 257
Query 252 SNGEALKGVNYTPLKAGE--ALNMQSLVSPVT-------SGISINDFRNVNAYQRYLELN 302
+G + + L G+ ++ Q+L+ P G++IND R NA QR+ E N
Sbjct 258 ISGSEVLYSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQMGVNINDIRTSNALQRWFERN 317
Query 303 QFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTG--AYVGAL 360
G Y E I F V L P++LGG I V+ + QT T +T A +
Sbjct 318 ARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGH 377
Query 361 GSQAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEF 420
G AG+ F + + +E ++G+M + P Y +PK + +D + PEF
Sbjct 378 GISAGVNHGF-------TRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEF 430
Query 421 DHIGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFIMYR 480
H+G Q I EL ++ N FGY + EY + HG F ++ + + R
Sbjct 431 AHLGEQEIKNEELYLNESDAANEG---TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNR 487
Query 481 SFVDVPELGKDFTTMKPGDVNNVFSVTEVG-DKILGQIHFDCTAQLPISRVVVPRL 535
F + P L F P N VF+ E DK QI+ D A + + P L
Sbjct 488 IFKEKPNLNTTFVECNPS--NRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 541
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 112 bits (281), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 113/402 (28%), Positives = 169/402 (42%), Gaps = 30/402 (7%)
Query 144 RAYEAVYNAYIRNTRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYANWQSDAYTSAL 203
RAY ++N + R+ + + + + G D+ T +L + D +TSAL
Sbjct 154 RAYNLIWNEWFRD----------ENLQDSVVVDKGDGPDTYTDYTLLRRGKRHDYFTSAL 203
Query 204 TAPQQGIAPLVGLTTYETKSINEAGHEVTV--VNTAIVDEDGNAFKVDFESNGEALKGVN 261
PQ+G A + L N+ G + V+T V + V E+NG V
Sbjct 204 PWPQKGDAVTLPLGGSANVVYNDTGDPAYIREVSTGNVWTTPSRESVSKEANGN--MSVP 261
Query 262 YTPLKAGEALNMQSLVSPVTSGI--SINDFRNVNAYQRYLELNQFRGFSYKEIIEGRFDV 319
+ A N SLV+ +++ +IN R QR LE + G Y EI+ F V
Sbjct 262 TGSVNAQYDPN-GSLVADLSTATAATINAIRQSFQIQRLLERDARGGTRYTEIVRSHFGV 320
Query 320 NVRYDALNMPEYLGGITRDIVVNPITQTVETSNTGAYVGALGSQAGLATCFGNTDGSISV 379
+ PEYLGG + I+VNP+ Q + +G LG+ + T + G S
Sbjct 321 ISPDARMQRPEYLGGGSAPIIVNPVAQQSASGASGTDT-PLGTLGAVGTGLASGHGFASS 379
Query 380 FCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYVRELAAVQAR 439
F E +V+G+ V Y L + + R D F P F H+G QPI +EL A
Sbjct 380 FT-EHGVVVGLCSVRADLTYQQGLHRMFSRSTRYDFFFPVFSHLGEQPILNKELYATGTS 438
Query 440 QFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLS----SLRNFIMYRSFVDVPELGKDFTTM 495
+ VFGYQ W EY KP + GL S +L + + ++F +P L F
Sbjct 439 TDD----DVFGYQEAWAEYRYKPSQVTGLMRSTAAGTLDAWHLAQNFGSLPTLNSTFIED 494
Query 496 KPGDVNNVFSVTEV--GDKILGQIHFDCTAQLPISRVVVPRL 535
P V+ V +V G + + FD P+ VP L
Sbjct 495 TP-PVDRVVAVGSEANGQQFIFDAFFDINMARPMPMYSVPGL 535
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 111 bits (277), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 121/414 (29%), Positives = 174/414 (42%), Gaps = 48/414 (12%)
Query 137 KISAYPFRAYEAVYNAYIRN---TRNNPFILNGKKTYNRFITNDAGGADSLTPKSLFYAN 193
++SA PFRAY+ +YN Y R+ T+ F LN + AD +T
Sbjct 158 QVSALPFRAYQLIYNEYYRDQNLTKPIEFSLNSGIVLS---------ADEVTRLLTLRRR 208
Query 194 -WQSDAYTSALTAPQQGIAPLVGLTTYETKSINEAGHEVTVVNTAIVDEDGNAFKVDFES 252
W+ D +TSAL Q+G P V T I +G + V T D + +++ S
Sbjct 209 TWEKDYFTSALPWVQRG--PEV------TVPIQGSGGNLDV--TLKNDAHADTYRMPGTS 258
Query 253 N--GEALKGVNYTPLKAG------EALNMQSLVSPVTSGISINDFRNVNAYQRYLELNQF 304
N A++ V + G E N Q V + G+SIND R NA QR+ E N
Sbjct 259 NRPAGAMQLVGGALIAGGTDGAYLEPDNFQVNVDEL--GVSINDLRTSNALQRWFERNAR 316
Query 305 RGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTG--AYVGALGS 362
G Y E I F V L P++LGG I V+ + QT T +T A + G
Sbjct 317 SGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSATDSTSPQANMAGHGI 376
Query 363 QAGLATCFGNTDGSISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEFDH 422
AG+ F + +E +IG+M + P Y +PK + +D + PEF H
Sbjct 377 SAGVNHGFKR-------YFEEHGYIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAH 429
Query 423 IGYQPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFIMYRSF 482
+G Q I E+ Q N FGY + EY + HG F ++ + + R F
Sbjct 430 LGEQEIKNEEVYLQQTPASN---NGTFGYTPRYAEYKYSMNEVHGDFRGNMAFWHLNRIF 486
Query 483 VDVPELGKDFTTMKPGDVNNVFSVTEVG-DKILGQIHFDCTAQLPISRVVVPRL 535
+ P L F P N VF+ E DK Q++ D A + + P L
Sbjct 487 SESPNLNTTFVECNPS--NRVFATAETSDDKYWIQLYQDVKALRLMPKYGTPML 538
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 105 bits (261), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 117/460 (25%), Positives = 179/460 (39%), Gaps = 74/460 (16%)
Query 112 DFSVTNYYAGVTPYYGVGSTDHKAI-------KISAYPFRAYEAVYNAYIRNTR-NNPFI 163
D+SV A + + DH I ++A PFRAY +YN + RN NP
Sbjct 107 DYSVPQVTAPAGGWEELSLADHMGIPTKVDNISVNALPFRAYGLIYNEFFRNQNLTNPTQ 166
Query 164 LN-------GKKTYNRFITNDAG--GADSLTPKSLFYANWQSDAYTSALTAPQQG----- 209
+ GK + +ND GA L F D +T AL PQ+G
Sbjct 167 VEVTDANIAGKNPNDVKNSNDWAITGAKCLKSAKFF------DYFTGALPQPQKGEPVEI 220
Query 210 --------------IAPLVGLTTYET---KSINEAGHEVTVVNTAIVDEDG----NAFKV 248
PL ++ +T +S + G+ +V ++G N K
Sbjct 221 NLASSWLPVGIGDYHGPLDKVSNSDTLTWESPSSEGNTKRTYALGMVQQEGEVNPNGLK- 279
Query 249 DFESNGEALKGVNYTPLKAGEALNMQSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFS 308
N E G +++ A A SPVT+ ++N R Q+ LE + G
Sbjct 280 ----NFETKAGGSFSESGAVAAYPTNLWASPVTAAATVNQLRQAFQVQKLLEKDARGGTR 335
Query 309 YKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTGAYVGALGSQAGLAT 368
Y+EI++ F V + +PEYLGG I V+ + QT +++ S G
Sbjct 336 YREILKNHFGVTTSDARMQIPEYLGGCKVPINVSQVVQTSASTD--------ASPQGNTA 387
Query 369 CFGNTDGSISVFC---DEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEFDHIGY 425
T S S+F DE +IGV Y + + + ++RLD + P +IG
Sbjct 388 AISVTPFSKSMFTKSFDEHGFIIGVATARTAQSYQQGIERMWSRKDRLDYYFPVLANIGE 447
Query 426 QPIYVRELAAVQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLS----SLRNFIMYRS 481
Q I +E+ A Q N FGYQ W +Y KP+ G F S SL + +
Sbjct 448 QAILNKEIYA----QGNAKDDEAFGYQEAWADYRYKPNTICGRFRSNAQQSLDAWHYGQD 503
Query 482 FVDVPELGKDFTTMKPGDVNNVFSVTEVGDKILGQIHFDC 521
+ +P L D+ ++ +V D + F+C
Sbjct 504 YDKLPTLSTDWMEQSDIEMKRTLAVQTEPD-FIANFRFNC 542
>gi|575096093|emb|CDL66973.1| unnamed protein product [uncultured bacterium]
Length=574
Score = 104 bits (260), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 111/457 (24%), Positives = 192/457 (42%), Gaps = 54/457 (12%)
Query 108 VGFDDFSVTNYYAGVTPYYGVGSTDH--------KAIKISAYPFRAYEAVYNAYIRNTRN 159
V +D+ S + + + Y+G+ S + + + +SA PFRAY ++N + R+
Sbjct 125 VRYDNLSKSTFIGTLVDYFGISSANFGSDKSGVTELVSVSALPFRAYWLIWNEWFRDENL 184
Query 160 NPFILNGKKTYNRFITNDAGGADSL--------------TPKSLFYANWQSDAYTSALTA 205
+ N + N G ++ P+ Y D +TS L
Sbjct 185 QSSVKVSMGDTNSAVDNMGSGTGNVNYSFPSGVTSYYHCAPRGKRY-----DYFTSCLPW 239
Query 206 PQQGIAPLVGLTTYETKSINEAGH-EVTVVNTAIVDEDGNAFKVDFESNGEALKGV---N 261
PQ+G P V L T +++ + +T+ + + G+ + + G+ L
Sbjct 240 PQKG--PGVELPLGSTANVSGQNNISLTLPSVYYNGDTGSGYSNLGQMVGKQLSSARQET 297
Query 262 YTPLKAGEALNMQSLVSPVT------SGISINDFRNVNAYQRYLELNQFRGFSYKEIIEG 315
Y+ +K L + +S ++ + I+IN R QRY E++ G Y E ++
Sbjct 298 YSYIKPAGNLTLNGSMSGLSVDLSSATSITINSLRQAFMLQRYYEVDARGGTRYTEKLQA 357
Query 316 RFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETSNTGAYVGALGSQAGLATCFGNTDG 375
F V L PEYLGG + +NP+ QT T++ + G+ A G T
Sbjct 358 HFGVTNPDSRLQRPEYLGGRSSMFNINPVAQTSSTND----ISPQGNMAAYG-IHGRTYR 412
Query 376 SISVFCDEESIVIGVMHVIPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIYVRELAA 435
+ + E +VIG+ V Y + ++ LD + PEF H+G Q + +E+
Sbjct 413 AFNKSFTEFGVVIGLCSVRADLTYQQGTERMWFRKDDLDFYWPEFAHLGEQAVLNQEI-Y 471
Query 436 VQARQFNVDPMTVFGYQRPWYEYVQKPDRAHGLFLSSLRNFI----MYRSFVDVPELGKD 491
VQ + VFGYQ + EY KP++ G F S+ + + + + F +P+LG
Sbjct 472 VQGTSADTG---VFGYQERYAEYRYKPNKITGQFRSTYKQTLDVWHLAQKFDSLPKLGDQ 528
Query 492 FTTMKPGDVNNVFSVTEVGDKILG-QIHFDCTAQLPI 527
F P V+ V +V +L + H C LP+
Sbjct 529 FIQDHP-PVSRVVAVPSYPHFLLDVKFHLQCVRPLPL 564
Lambda K H a alpha
0.319 0.136 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3854736288630