bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_1
Length=598
Score E
Sequences producing significant alignments: (Bits) Value
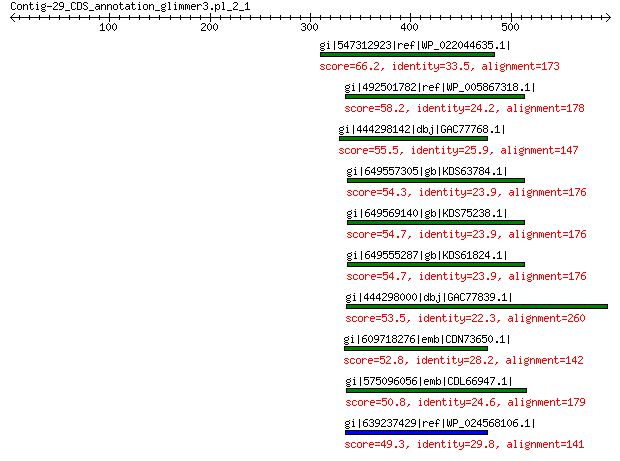
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 66.2 2e-08
gi|492501782|ref|WP_005867318.1| hypothetical protein 58.2 1e-05
gi|444298142|dbj|GAC77768.1| major capsid protein 55.5 4e-05
gi|649557305|gb|KDS63784.1| capsid family protein 54.3 6e-05
gi|649569140|gb|KDS75238.1| capsid family protein 54.7 1e-04
gi|649555287|gb|KDS61824.1| capsid family protein 54.7 1e-04
gi|444298000|dbj|GAC77839.1| major capsid protein 53.5 3e-04
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 52.8 5e-04
gi|575096056|emb|CDL66947.1| unnamed protein product 50.8 0.002
gi|639237429|ref|WP_024568106.1| hypothetical protein 49.3 0.007
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 66.2 bits (160), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 58/193 (30%), Positives = 86/193 (45%), Gaps = 23/193 (12%)
Query 310 VPSNPDRFSRLVPVGSSSAVSM-----------TGVTT-IPQLAIASRLQEYKDLLGAGG 357
VP +PD F ++ GSS AV + TG + +P+L + +++Q + D L G
Sbjct 15 VPYSPDLFGNIIKQGSSPAVEIEVMNALDLNISTGFSVAVPELRLRTKIQNWMDRLFVSG 74
Query 358 SRYSDWLETFFASKIE--HVDRPKLLFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGGSI 415
R D T + +K +V++P L ++N V A + + LGQ +
Sbjct 75 GRVGDVFRTLWGTKSSAIYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACV 134
Query 416 A----FNERLGRRQSYYFREPG--YLIDMLSIRPVYYWSFIKPDYLNYSGSDYFNPIYND 469
F+ G YY +EPG LI ML P Y + PD + S D FNP N
Sbjct 135 DRYCDFSGHSGI--DYYAKEPGTFMLITMLVPEPAYSQG-LHPDLASISFGDDFNPELNG 191
Query 470 IGYQDVPAFRIAF 482
IG+Q VP R +
Sbjct 192 IGFQLVPRHRFSM 204
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 58.2 bits (139), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 43/184 (23%), Positives = 82/184 (45%), Gaps = 10/184 (5%)
Query 335 TTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFA--SKIEHVDRPKLLFSASQTVNVQI 392
+I L ++ LQ + + GSRY + + + F S + RP+ L ++V
Sbjct 296 VSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 355
Query 393 VMNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIK 451
V+ + ++ S + G S N R YF E GY+I ++SIRP Y +
Sbjct 356 VLQTSATDSTSPQANMAGHGISAGVNHGFKR----YFEEHGYIIGIMSIRPRTGYQQGVP 411
Query 452 PDYLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSAS---EPCFNEFRSSYDEVLG 508
D+ + D++ P + +G Q++ + P +++ + P + E++ S +EV G
Sbjct 412 KDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNNGTFGYTPRYAEYKYSMNEVHG 471
Query 509 QLQA 512
+
Sbjct 472 DFRG 475
>gi|444298142|dbj|GAC77768.1| major capsid protein [uncultured marine virus]
Length=299
Score = 55.5 bits (132), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 38/149 (26%), Positives = 69/149 (46%), Gaps = 4/149 (3%)
Query 329 VSMTGVTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETF-FASKIEHVDRPKLLFSASQT 387
+S G I L A LQ Y++ G+R++++L +S + RP+++ +
Sbjct 113 LSQAGAININDLREAFALQRYQEARNLYGARFTEYLRYLGISSSXGRLQRPEMISTGKSN 172
Query 388 VNVQIVMNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYY 446
+N V+N G + + PLG+ GG + R Y+ E G++I ++S+RP Y
Sbjct 173 INFSEVLNTTGPSGVD-DHPLGEMGGHGIAGVK-SNRARYFCEEHGHIISLMSVRPKTIY 230
Query 447 WSFIKPDYLNYSGSDYFNPIYNDIGYQDV 475
+ + S DY+ IG ++V
Sbjct 231 MTTQHKQFDRESKEDYWQKELQAIGMEEV 259
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 54.3 bits (129), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 42/182 (23%), Positives = 81/182 (45%), Gaps = 10/182 (5%)
Query 337 IPQLAIASRLQEYKDLLGAGGSRYSDWLETFFA--SKIEHVDRPKLLFSASQTVNVQIVM 394
I + ++ LQ + + GSRY + + + F S + RP+ L ++V V+
Sbjct 5 INDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVL 64
Query 395 NQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIKPD 453
+ ++ S + G S N R YF E GY++ ++SIRP Y + D
Sbjct 65 QTSSTDSTSPQANMAGHGISAGVNHGFTR----YFEEHGYIMGIMSIRPRTGYQQGVPKD 120
Query 454 YLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSAS---EPCFNEFRSSYDEVLGQL 510
+ + D++ P + +G Q++ + N + A+ + P + E++ S +EV G
Sbjct 121 FRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDF 180
Query 511 QA 512
+
Sbjct 181 RG 182
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 54.7 bits (130), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/182 (23%), Positives = 81/182 (45%), Gaps = 10/182 (5%)
Query 337 IPQLAIASRLQEYKDLLGAGGSRYSDWLETFFA--SKIEHVDRPKLLFSASQTVNVQIVM 394
I + ++ LQ + + GSRY + + + F S + RP+ L ++V V+
Sbjct 150 INDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVL 209
Query 395 NQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIKPD 453
+ ++ S + G S N R YF E GY++ ++SIRP Y + D
Sbjct 210 QTSSTDSTSPQANMAGHGISAGVNHGFTR----YFEEHGYIMGIMSIRPRTGYQQGVPKD 265
Query 454 YLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSAS---EPCFNEFRSSYDEVLGQL 510
+ + D++ P + +G Q++ + N + A+ + P + E++ S +EV G
Sbjct 266 FRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDF 325
Query 511 QA 512
+
Sbjct 326 RG 327
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 54.7 bits (130), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/182 (23%), Positives = 81/182 (45%), Gaps = 10/182 (5%)
Query 337 IPQLAIASRLQEYKDLLGAGGSRYSDWLETFFA--SKIEHVDRPKLLFSASQTVNVQIVM 394
I + ++ LQ + + GSRY + + + F S + RP+ L ++V V+
Sbjct 301 INDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVL 360
Query 395 NQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIKPD 453
+ ++ S + G S N R YF E GY++ ++SIRP Y + D
Sbjct 361 QTSSTDSTSPQANMAGHGISAGVNHGFTR----YFEEHGYIMGIMSIRPRTGYQQGVPKD 416
Query 454 YLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSAS---EPCFNEFRSSYDEVLGQL 510
+ + D++ P + +G Q++ + N + A+ + P + E++ S +EV G
Sbjct 417 FRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYTPRYAEYKYSQNEVHGDF 476
Query 511 QA 512
+
Sbjct 477 RG 478
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 53.5 bits (127), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 58/269 (22%), Positives = 106/269 (39%), Gaps = 35/269 (13%)
Query 336 TIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFAS-KIEHVDRPKLLFSASQTVNVQIVM 394
TI + A +Q Y++ GSRY+++L + K + RP+ + + +N V+
Sbjct 237 TINDIRRAFAIQRYQEARSRYGSRYTEYLRYLGVNPKDARLQRPEYMGGGTTQINFSEVL 296
Query 395 NQAGDNNFSGNQPLGQQGGSIAFNERLGRRQS----YYFREPGYLIDMLSIRP-VYYWSF 449
+ + G + Q G + + +S Y E GY+I MLS+RP Y +
Sbjct 297 QTSPE--IPGEDQVSQFGVGDMYGHGIAAMRSNKYRRYIEEHGYIISMLSVRPKTMYTNG 354
Query 450 IKPDYLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSASEPCFNEFRSSYDEVLGQ 509
I +L + DY+ IG Q++ I + G + +N+ S Y E
Sbjct 355 IHRSWLRLTKEDYYQKELEHIGQQEIMNNEIYADEGAGTETFG---YNDRYSEYRETPSH 411
Query 510 LQAYYRSPAEGGTGAPLYSYWVQQRAVLTSSSTGSLPESYYYPVL---FTDLSQVNSPFS 566
+ A +R + +YW R E PVL F D +
Sbjct 412 VSAEFRG---------ILNYWHMAR------------EFEAPPVLNQSFVDCDATKRIHN 450
Query 567 STVEDNFFVNLSYAVQKKNLVNKTFATRL 595
+D ++ + + + + L+++ A R+
Sbjct 451 EQTQDALWIMIQHKMVARRLLSRNAAPRI 479
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 52.8 bits (125), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 40/145 (28%), Positives = 69/145 (48%), Gaps = 7/145 (5%)
Query 334 VTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASKIE--HVDRPKLLFSASQTVNVQ 391
V+T+ L A +LQE+ + GSRY++ + +FF K + RP+ L + +
Sbjct 288 VSTVNDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKSPIMIS 347
Query 392 IVMNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRPVYYWSFIK 451
V+ Q+ ++ + + G I + R +F E GY+I ++S+ P +S
Sbjct 348 EVLQQSATDSTTPQGNMAGHGIGIGKDGGFSR----FFEEHGYVIGLMSVIPKTSYSQGI 403
Query 452 PDYLNYSGS-DYFNPIYNDIGYQDV 475
P + + S DYF P + IG Q V
Sbjct 404 PRHFSKSDKFDYFWPQFEHIGEQPV 428
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 50.8 bits (120), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 44/186 (24%), Positives = 85/186 (46%), Gaps = 11/186 (6%)
Query 336 TIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFA--SKIEHVDRPKLLFSASQTVNVQIV 393
TI QL +A ++Q++ + GGSRY++ + +FF S + R + L +N+ V
Sbjct 318 TINQLRMAFQIQKFYEKQARGGSRYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQV 377
Query 394 MNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYY--FREPGYLIDMLSIRPVY-YWSFI 450
+ Q+G +G+ QG + ++ + F E G++I ++ R + Y I
Sbjct 378 IQQSG----TGSASTTPQGTVVGMSQTTDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGI 433
Query 451 KPDYLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASS--ASEPCFNEFRSSYDEVLG 508
+ DY+ P++++IG Q + I GN + + E+R V G
Sbjct 434 DRMWSRKDKFDYYWPVFSNIGEQAIKNKEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTG 493
Query 509 QLQAYY 514
++++ Y
Sbjct 494 EMRSSY 499
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 49.3 bits (116), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 42/145 (29%), Positives = 69/145 (48%), Gaps = 9/145 (6%)
Query 335 TTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASKIE--HVDRPKLLFSASQTVNVQI 392
+TI L A +LQE+ + GSRY++ + +FF K + RP+ L + +
Sbjct 298 STINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDGRLQRPEFLGGNKTPILISE 357
Query 393 VMNQAGDNNFSGNQPLGQQGG-SIAFNERLGRRQSYYFREPGYLIDMLSIRPVYYWSFIK 451
V+ Q+ ++ P G G I+ + G S +F E GY+I ++S+ P +S
Sbjct 358 VLQQSSTDS---TTPQGNMAGHGISVGKEGGF--SKFFEEHGYVIGLMSVIPKTSYSQGI 412
Query 452 PDYL-NYSGSDYFNPIYNDIGYQDV 475
P + + DYF P + IG Q V
Sbjct 413 PRHFSKFDKFDYFWPQFEHIGEQPV 437
Lambda K H a alpha
0.318 0.134 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4446915032268