bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-40_CDS_annotation_glimmer3.pl_2_1
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
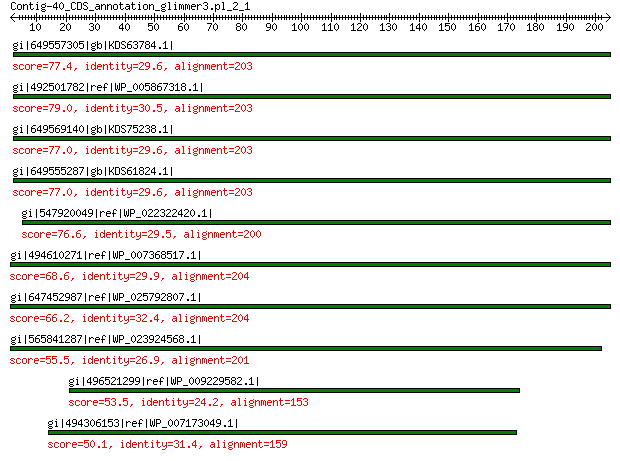
gi|649557305|gb|KDS63784.1| capsid family protein 77.4 5e-14
gi|492501782|ref|WP_005867318.1| hypothetical protein 79.0 9e-14
gi|649569140|gb|KDS75238.1| capsid family protein 77.0 2e-13
gi|649555287|gb|KDS61824.1| capsid family protein 77.0 5e-13
gi|547920049|ref|WP_022322420.1| capsid protein VP1 76.6 6e-13
gi|494610271|ref|WP_007368517.1| capsid protein 68.6 3e-10
gi|647452987|ref|WP_025792807.1| hypothetical protein 66.2 2e-09
gi|565841287|ref|WP_023924568.1| hypothetical protein 55.5 5e-06
gi|496521299|ref|WP_009229582.1| capsid protein 53.5 2e-05
gi|494306153|ref|WP_007173049.1| hypothetical protein 50.1 3e-04
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 77.4 bits (189), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 60/205 (29%), Positives = 98/205 (48%), Gaps = 16/205 (8%)
Query 2 QSEIAFDEIVSNSATEE-EPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRID 60
++ I+ E++ S+T+ P +AG G++ G E IM + SI PR
Sbjct 55 RTPISVSEVLQTSSTDSTSPQANMAGHGISA--GVNHGFTRYFEEHGYIMGIMSIRPRTG 112
Query 61 YSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSW 119
Y QG K + + NMD F+ P +G QE+ EE ++A ++ + G P +
Sbjct 113 YQQGVPKDFRKFDNMD-FYFPEFAHLGEQEIKNEELYLNESDAANE-----GTFGYTPRY 166
Query 120 IEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFAESRLSS 179
EY NE +G+F M AF LNR+++E + +T+++ N +FA + S
Sbjct 167 AEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRVFATAETSD 220
Query 180 QNFWVQVAFDVTARRVMSAKQIPNL 204
+WVQ+ D+ A R+M P L
Sbjct 221 DKYWVQIYQDIKALRLMPKYGTPML 245
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 79.0 bits (193), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 62/205 (30%), Positives = 100/205 (49%), Gaps = 16/205 (8%)
Query 2 QSEIAFDEIVSNSATEE-EPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRID 60
++ I+ E++ SAT+ P +AG G++ G K E I+ + SI PR
Sbjct 348 RTPISVSEVLQTSATDSTSPQANMAGHGISA--GVNHGFKRYFEEHGYIIGIMSIRPRTG 405
Query 61 YSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSW 119
Y QG K + + NMD F+ P +G QE+ EE T A+++ + G P +
Sbjct 406 YQQGVPKDFRKFDNMD-FYFPEFAHLGEQEIKNEEVYLQQTPASNN-----GTFGYTPRY 459
Query 120 IEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFAESRLSS 179
EY +NE +G+F M AF LNR++ E+ + +T+++ N +FA + S
Sbjct 460 AEYKYSMNEVHGDFRGNM--AFWHLNRIFSESPNLN----TTFVECNPSNRVFATAETSD 513
Query 180 QNFWVQVAFDVTARRVMSAKQIPNL 204
+W+Q+ DV A R+M P L
Sbjct 514 DKYWIQLYQDVKALRLMPKYGTPML 538
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 77.0 bits (188), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 60/205 (29%), Positives = 98/205 (48%), Gaps = 16/205 (8%)
Query 2 QSEIAFDEIVSNSATEE-EPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRID 60
++ I+ E++ S+T+ P +AG G++ G E IM + SI PR
Sbjct 200 RTPISVSEVLQTSSTDSTSPQANMAGHGISA--GVNHGFTRYFEEHGYIMGIMSIRPRTG 257
Query 61 YSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSW 119
Y QG K + + NMD F+ P +G QE+ EE ++A ++ + G P +
Sbjct 258 YQQGVPKDFRKFDNMD-FYFPEFAHLGEQEIKNEELYLNESDAANE-----GTFGYTPRY 311
Query 120 IEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFAESRLSS 179
EY NE +G+F M AF LNR+++E + +T+++ N +FA + S
Sbjct 312 AEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRVFATAETSD 365
Query 180 QNFWVQVAFDVTARRVMSAKQIPNL 204
+WVQ+ D+ A R+M P L
Sbjct 366 DKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 77.0 bits (188), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 60/205 (29%), Positives = 98/205 (48%), Gaps = 16/205 (8%)
Query 2 QSEIAFDEIVSNSATEE-EPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRID 60
++ I+ E++ S+T+ P +AG G++ G E IM + SI PR
Sbjct 351 RTPISVSEVLQTSSTDSTSPQANMAGHGISA--GVNHGFTRYFEEHGYIMGIMSIRPRTG 408
Query 61 YSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSW 119
Y QG K + + NMD F+ P +G QE+ EE ++A ++ + G P +
Sbjct 409 YQQGVPKDFRKFDNMD-FYFPEFAHLGEQEIKNEELYLNESDAANE-----GTFGYTPRY 462
Query 120 IEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFAESRLSS 179
EY NE +G+F M AF LNR+++E + +T+++ N +FA + S
Sbjct 463 AEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRVFATAETSD 516
Query 180 QNFWVQVAFDVTARRVMSAKQIPNL 204
+WVQ+ D+ A R+M P L
Sbjct 517 DKYWVQIYQDIKALRLMPKYGTPML 541
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 76.6 bits (187), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 59/202 (29%), Positives = 95/202 (47%), Gaps = 16/202 (8%)
Query 5 IAFDEIVSNSATEE-EPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRIDYSQ 63
I+ E++ S+T+E P +AG G++ +G K E I+ + SITPR Y Q
Sbjct 366 ISVSEVLQTSSTDETSPQANMAGHGISAGINNG--FKHYFEEHGYIIGIMSITPRSGYQQ 423
Query 64 G-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSWIEY 122
G + +T+ NMD F+ P + QE+ ++D + G P + EY
Sbjct 424 GVPRDFTKFDNMD-FYFPEFAHLSEQEI-----KNQELFVSEDAAYNNGTFGYTPRYAEY 477
Query 123 TTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFAESRLSSQNF 182
+E +G+F L+F LNR++E+ + +T+++ N +FA S F
Sbjct 478 KYHPSEAHGDFRGN--LSFWHLNRIFEDKPNLN----TTFVECKPSNRVFATSETEDDKF 531
Query 183 WVQVAFDVTARRVMSAKQIPNL 204
WVQ+ DV A R+M P L
Sbjct 532 WVQMYQDVKALRLMPKYGTPML 553
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 68.6 bits (166), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 61/238 (26%), Positives = 101/238 (42%), Gaps = 38/238 (16%)
Query 1 MQSEIAFDEIVSNS-----ATEEEPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSI 55
+ + E+V+ S A E LG L G+GV ++ S +K E +IM + S+
Sbjct 298 FDNPVVISEVVNQSEFDRGADESPCLGDLGGKGVGSLNSSSIDFDVK--EHGIIMCIYSV 355
Query 56 TPRIDYSQGNKW--WTRLQNMDDFHKPTLDAIGFQELIae-----------------eaa 96
P+ +Y G + + R +DF +P +G+Q ++ +
Sbjct 356 VPQTEY-NGTYFDPFNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDNPVPDGPEKQKR 414
Query 97 awtteatDDHELVYQSLGKQPSWIEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTI 156
E + LG Q + EY T + +GEF +G+ L++ C R Y+ D
Sbjct 415 LAAGYPLSSIEANNRLLGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSPR-YDFGFDGKA 473
Query 157 GN----------ASTYIDPTIYNSIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 204
G+ A Y++P+I N+IF S + + +F V FDV A R MS + L
Sbjct 474 GDKKLVNSPWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDVKAVRPMSVSGLAGL 531
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 66.2 bits (160), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 66/241 (27%), Positives = 105/241 (44%), Gaps = 43/241 (18%)
Query 1 MQSEIAFDEIVS---NSATE--EEPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSI 55
+ I E+VS N+A++ +G L G+G+ +M S ++ TE +IM + S+
Sbjct 350 FDNSIVVSEVVSTNGNAASDGSHASIGDLGGKGIGSM--SSGTIEFDSTEHGIIMCIYSV 407
Query 56 TPRIDYSQG-----NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatD------ 104
P+ +Y+ N+ TR Q F++P +G+Q LI + T +
Sbjct 408 APQSEYNASYLDPFNRKLTREQ----FYQPEFADLGYQALIGSDLICSTLGMNEKQAGFS 463
Query 105 DHELVYQSLGKQPSWIEYTTDVNETYGEFAAGMPLAFMCLNR-----------VYEENSD 153
D EL LG Q + EY T + +G+F +G L++ C R + EN
Sbjct 464 DIELNNNLLGYQVRYNEYKTARDLVFGDFESGKSLSYWCTPRFDFGYGDTEKKIAPENKG 523
Query 154 ----HTIGNAST------YIDPTIYNSIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPN 203
GN S YI+P + N IF S + + +F V DV A R MS + +
Sbjct 524 GADYRKKGNRSHWSSRNFYINPNLVNPIFLTSAVQADHFIVNSFLDVKAVRPMSVTGLSS 583
Query 204 L 204
L
Sbjct 584 L 584
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 55.5 bits (132), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 54/216 (25%), Positives = 94/216 (44%), Gaps = 20/216 (9%)
Query 1 MQSEIAFDEIVSNS-------ATEEEPLGTLAGRGVATMYKSGRGLKIKCTEPSMIMALG 53
++I+ E+V+ S A+ +G + G+G+ M SG + E +IM +
Sbjct 443 FDNQISISEVVTTSNGSVDGTASTGSVVGQVFGKGIGAM-NSGH-ISYDVKEHGLIMCIY 500
Query 54 SITPRIDY-SQGNKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawtteatDDHELVYQS 112
SI P++DY ++ + R + +D+ +P + +G Q +I + A D + +
Sbjct 501 SIAPQVDYDARELDPFNRKFSREDYFQPEFENLGMQPVIQSDLCLCINSAKSDSSDQHNN 560
Query 113 -LGKQPSWIEYTTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNAS---TYIDPTIY 168
LG ++EY T + +GEF +G L+ + N G S +DP +
Sbjct 561 VLGYSARYLEYKTARDIIFGEFMSGGSLSAWATPK---NNYTFEFGKLSLPDLLVDPKVL 617
Query 169 NSIFA---ESRLSSQNFWVQVAFDVTARRVMSAKQI 201
IFA +S+ F V FDV A R M +
Sbjct 618 EPIFAVKYNGSMSTDQFLVNSYFDVKAIRPMQVNDM 653
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 53.5 bits (127), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/154 (24%), Positives = 69/154 (45%), Gaps = 12/154 (8%)
Query 21 LGTLAGRGVATMYKSGRGLKIKCTEPSMIMALGSITPRIDYSQGN-KWWTRLQNMDDFHK 79
LG + G+G + Y ++ EP ++M + S+ P + Y + Q D+
Sbjct 365 LGKITGKGTGSGYGE---IQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFI 421
Query 80 PTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSWIEYTTDVNETYGEFAAGMPL 139
P + +G Q ++ + + S G QP + EY T + +G+FA G PL
Sbjct 422 PEFENLGMQPIVPAFVSLNRAKD--------NSYGWQPRYSEYKTAFDINHGQFANGEPL 473
Query 140 AFMCLNRVYEENSDHTIGNASTYIDPTIYNSIFA 173
++ + R ++ +T A+ I+P +S+FA
Sbjct 474 SYWSIARARGSDTLNTFNVAALKINPHWLDSVFA 507
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 50.1 bits (118), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 50/170 (29%), Positives = 75/170 (44%), Gaps = 25/170 (15%)
Query 14 SATEEEP----LGTLAGRGVATMYKSGRG-LKIKCTEPSMIMALGSITPRIDYSQGNKWW 68
+ATE +P LG +AG+G SGRG + E ++M + S+ P+I Y
Sbjct 329 TATEYKPEAGYLGRIAGKGTG----SGRGRIVFDAKEHGVLMCIYSLVPQIQYD-----C 379
Query 69 TRLQNMDD------FHKPTLDAIGFQELIaeeaaawtteatDDHELVYQSLGKQPSWIEY 122
TRL M D F P + +G Q L + + D V LG QP + EY
Sbjct 380 TRLDPMVDKLDRFDFFTPEFENLGMQPL--NSSYISSFCTPDPKNPV---LGYQPRYSEY 434
Query 123 TTDVNETYGEFAAGMPLAFMCLNRVYEENSDHTIGNASTYIDPTIYNSIF 172
T ++ +G+FA L+ ++R + + A IDP NS+F
Sbjct 435 KTALDINHGQFAQNDALSSWSVSRFRRWTTFPQLEIADFKIDPGCLNSVF 484
Lambda K H a alpha
0.317 0.132 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 671121370458