bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-41_CDS_annotation_glimmer3.pl_2_1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
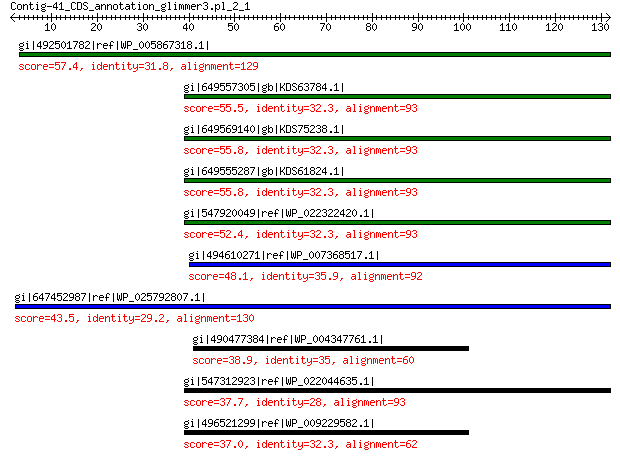
gi|492501782|ref|WP_005867318.1| hypothetical protein 57.4 4e-07
gi|649557305|gb|KDS63784.1| capsid family protein 55.5 5e-07
gi|649569140|gb|KDS75238.1| capsid family protein 55.8 1e-06
gi|649555287|gb|KDS61824.1| capsid family protein 55.8 2e-06
gi|547920049|ref|WP_022322420.1| capsid protein VP1 52.4 2e-05
gi|494610271|ref|WP_007368517.1| capsid protein 48.1 4e-04
gi|647452987|ref|WP_025792807.1| hypothetical protein 43.5 0.013
gi|490477384|ref|WP_004347761.1| capsid protein 38.9 0.45
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 37.7 1.2
gi|496521299|ref|WP_009229582.1| capsid protein 37.0 2.2
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 57.4 bits (137), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 41/129 (32%), Positives = 64/129 (50%), Gaps = 11/129 (9%)
Query 3 DFHKPTLDAIGFQELIaeeaaawsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAA 62
DF+ P +G QE+ EE T A+ N + G P + EY +NE +GDF
Sbjct 421 DFYFPEFAHLGEQEIKNEEVYLQQTPASNNG-----TFGYTPRYAEYKYSMNEVHGDFRG 475
Query 63 GMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRV 122
M AF LNR++ E+ + +T+++ N++FA + S +W+Q+ DV A R+
Sbjct 476 NM--AFWHLNRIFSESPNLN----TTFVECNPSNRVFATAETSDDKYWIQLYQDVKALRL 529
Query 123 MSAKQIPNL 131
M P L
Sbjct 530 MPKYGTPML 538
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 55.5 bits (132), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 48/93 (52%), Gaps = 6/93 (6%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKI 98
+ G P + EY NE +GDF M AF LNR+++E + +T+++ N++
Sbjct 159 TFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRV 212
Query 99 FAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
FA + S +WVQ+ D+ A R+M P L
Sbjct 213 FATAETSDDKYWVQIYQDIKALRLMPKYGTPML 245
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 55.8 bits (133), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 48/93 (52%), Gaps = 6/93 (6%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKI 98
+ G P + EY NE +GDF M AF LNR+++E + +T+++ N++
Sbjct 304 TFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRV 357
Query 99 FAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
FA + S +WVQ+ D+ A R+M P L
Sbjct 358 FATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 55.8 bits (133), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 48/93 (52%), Gaps = 6/93 (6%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKI 98
+ G P + EY NE +GDF M AF LNR+++E + +T+++ N++
Sbjct 455 TFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFVECNPSNRV 508
Query 99 FAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
FA + S +WVQ+ D+ A R+M P L
Sbjct 509 FATAETSDDKYWVQIYQDIKALRLMPKYGTPML 541
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 52.4 bits (124), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 47/93 (51%), Gaps = 6/93 (6%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKI 98
+ G P + EY +E +GDF L+F LNR++E+ + +T+++ N++
Sbjct 467 TFGYTPRYAEYKYHPSEAHGDFRGN--LSFWHLNRIFEDKPNLN----TTFVECKPSNRV 520
Query 99 FAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
FA S FWVQ+ DV A R+M P L
Sbjct 521 FATSETEDDKFWVQMYQDVKALRLMPKYGTPML 553
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 48.1 bits (113), Expect = 4e-04, Method: Composition-based stats.
Identities = 33/102 (32%), Positives = 51/102 (50%), Gaps = 11/102 (11%)
Query 40 LGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYE-----ENSDHTIAN-----ASTY 89
LG Q + EY T + +G+F +G+ L++ C R Y+ + D + N A Y
Sbjct 431 LGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSPR-YDFGFDGKAGDKKLVNSPWSPAHFY 489
Query 90 IDPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
++P+I N IF S + + +F V FDV A R MS + L
Sbjct 490 VNPSILNTIFLVSAVKADHFLVNSFFDVKAVRPMSVSGLAGL 531
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 43.5 bits (101), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 38/157 (24%), Positives = 64/157 (41%), Gaps = 27/157 (17%)
Query 2 DDFHKPTLDAIGFQELI------aeeaaawsteatGNHELVYQSLGKQPSWIEYTTDVNE 55
+ F++P +G+Q LI + + EL LG Q + EY T +
Sbjct 428 EQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDIELNNNLLGYQVRYNEYKTARDL 487
Query 56 TYGDFAAGMPLAFMCLNRV--------------------YEENSDHT-IANASTYIDPTI 94
+GDF +G L++ C R Y + + + ++ + YI+P +
Sbjct 488 VFGDFESGKSLSYWCTPRFDFGYGDTEKKIAPENKGGADYRKKGNRSHWSSRNFYINPNL 547
Query 95 YNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
N IF S + + +F V DV A R MS + +L
Sbjct 548 VNPIFLTSAVQADHFIVNSFLDVKAVRPMSVTGLSSL 584
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 38.9 bits (89), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (52%), Gaps = 0/60 (0%)
Query 41 GKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKIFA 100
G QP + EY T ++ +G FA G PL++ + R + T AS I+P + IFA
Sbjct 459 GWQPRYSEYKTSLDINHGQFANGQPLSYWTVGRGRAGETLETFDIASLKINPKWLDSIFA 518
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 37.7 bits (86), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 46/104 (44%), Gaps = 15/104 (14%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYE-----------ENSDHTIANAS 87
S+G++ +W TD + +GDFA + L R + ++ ++T
Sbjct 237 SVGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYT----G 292
Query 88 TYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 131
TYI+P + +F + L + NF FD+ +SA +P L
Sbjct 293 TYINPLDWQYVFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 37.0 bits (84), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 39 SLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNKI 98
S G QP + EY T + +G FA G PL++ + R ++ +T A+ I+P + +
Sbjct 446 SYGWQPRYSEYKTAFDINHGQFANGEPLSYWSIARARGSDTLNTFNVAALKINPHWLDSV 505
Query 99 FA 100
FA
Sbjct 506 FA 507
Lambda K H a alpha
0.319 0.133 0.401 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 435513260292