bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_8
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
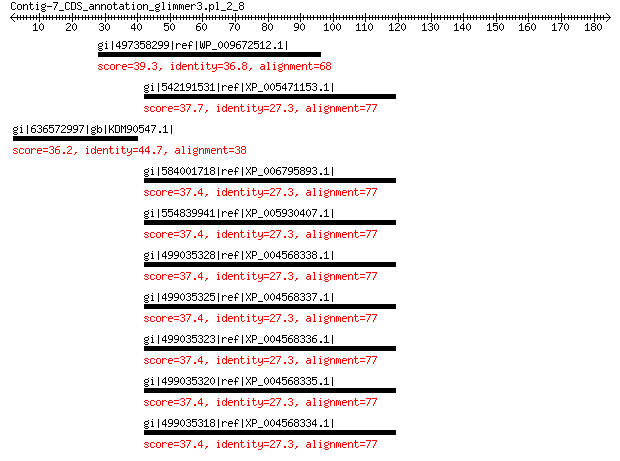
gi|497358299|ref|WP_009672512.1| MULTISPECIES: bacitracin synthe... 39.3 1.3
gi|542191531|ref|XP_005471153.1| PREDICTED: protein-methionine s... 37.7 4.1
gi|636572997|gb|KDM90547.1| hypothetical protein EA58_16625 36.2 5.1
gi|584001718|ref|XP_006795893.1| PREDICTED: protein-methionine s... 37.4 5.6
gi|554839941|ref|XP_005930407.1| PREDICTED: protein-methionine s... 37.4 5.8
gi|499035328|ref|XP_004568338.1| PREDICTED: protein-methionine s... 37.4 5.8
gi|499035325|ref|XP_004568337.1| PREDICTED: protein-methionine s... 37.4 5.8
gi|499035323|ref|XP_004568336.1| PREDICTED: protein-methionine s... 37.4 5.8
gi|499035320|ref|XP_004568335.1| PREDICTED: protein-methionine s... 37.4 5.8
gi|499035318|ref|XP_004568334.1| PREDICTED: protein-methionine s... 37.4 5.8
>gi|497358299|ref|WP_009672512.1| MULTISPECIES: bacitracin synthetase 1 [Paenibacillus]
gi|333607855|gb|EGL19165.1| AMP-binding enzyme [Paenibacillus sp. HGF7]
gi|512138639|gb|EPD81222.1| amino acid adenylation domain-containing protein [Paenibacillus
sp. HGH0039]
Length=2704
Score = 39.3 bits (90), Expect = 1.3, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 35/69 (51%), Gaps = 2/69 (3%)
Query 28 PEALRFIYNACRLSLYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRAF 87
P + I + LY EL G SA+YR L NL +H++ +P +HI F G
Sbjct 2370 PSVQQRIMSGLLKELYFRPYELGG-SALYRFALVSFNLREHYLLLPCSHIIFDGMSSEIL 2428
Query 88 RT-IYAFYS 95
RT + AFY+
Sbjct 2429 RTDVLAFYN 2437
>gi|542191531|ref|XP_005471153.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
[Oreochromis niloticus]
Length=2532
Score = 37.7 bits (86), Expect = 4.1, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRVFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|636572997|gb|KDM90547.1| hypothetical protein EA58_16625 [Photobacterium halotolerans]
Length=138
Score = 36.2 bits (82), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/38 (45%), Positives = 22/38 (58%), Gaps = 0/38 (0%)
Query 2 TLFEQAKRTFFVIKKYLQVDHSLDNAPEALRFIYNACR 39
+ FE T+FV+KK LQ N+PE L+ Y ACR
Sbjct 15 SFFEGGVNTYFVMKKGLQYSAREKNSPEDLKGQYEACR 52
>gi|584001718|ref|XP_006795893.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
[Neolamprologus brichardi]
Length=1271
Score = 37.4 bits (85), Expect = 5.6, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|554839941|ref|XP_005930407.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X2 [Haplochromis burtoni]
Length=2000
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|499035328|ref|XP_004568338.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X7 [Maylandia zebra]
Length=2016
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|499035325|ref|XP_004568337.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X6 [Maylandia zebra]
Length=2033
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|499035323|ref|XP_004568336.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X5 [Maylandia zebra]
Length=2041
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|499035320|ref|XP_004568335.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X4 [Maylandia zebra]
Length=2050
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
>gi|499035318|ref|XP_004568334.1| PREDICTED: protein-methionine sulfoxide oxidase mical3a-like
isoform X3 [Maylandia zebra]
Length=2058
Score = 37.4 bits (85), Expect = 5.8, Method: Composition-based stats.
Identities = 21/79 (27%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 42 LYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRRA--FRTIYAFYSYMDS 99
L+ NF + C + + A++ LCDH PS+H FY +L+ + A ++ +D
Sbjct 20 LFDNFVQATTCKGILK---AFQELCDHLEVKPSDHRIFYHKLKSKLNYWKAKALWAKLDK 76
Query 100 KHLHDQLLKVRSWSQNNYL 118
+ H + K R+ + L
Sbjct 77 RASHKEYKKGRACANTKCL 95
Lambda K H a alpha
0.329 0.139 0.439 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 496788675030