bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_1
Length=461
Score E
Sequences producing significant alignments: (Bits) Value
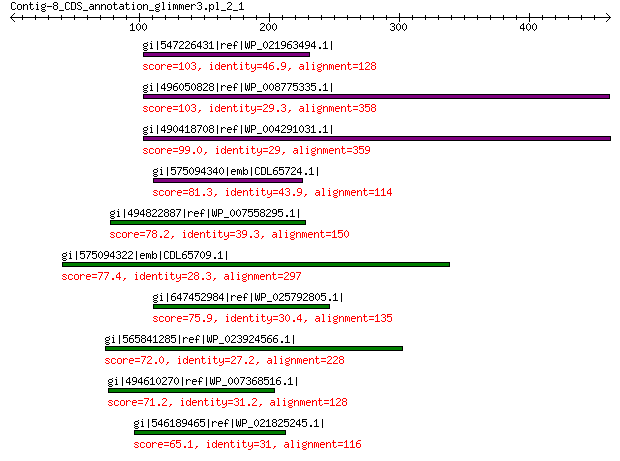
gi|547226431|ref|WP_021963494.1| predicted protein 103 1e-20
gi|496050828|ref|WP_008775335.1| hypothetical protein 103 2e-20
gi|490418708|ref|WP_004291031.1| hypothetical protein 99.0 3e-19
gi|575094340|emb|CDL65724.1| unnamed protein product 81.3 3e-13
gi|494822887|ref|WP_007558295.1| hypothetical protein 78.2 4e-12
gi|575094322|emb|CDL65709.1| unnamed protein product 77.4 6e-12
gi|647452984|ref|WP_025792805.1| hypothetical protein 75.9 2e-11
gi|565841285|ref|WP_023924566.1| hypothetical protein 72.0 3e-10
gi|494610270|ref|WP_007368516.1| hypothetical protein 71.2 4e-10
gi|546189465|ref|WP_021825245.1| hypothetical protein 65.1 5e-08
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 103 bits (258), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 60/135 (44%), Positives = 78/135 (58%), Gaps = 10/135 (7%)
Query 103 GLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEI 162
G L Y + RD QLF KR+RK LSK EKI Y+VSEY PKTFR H+H+LFF+D +
Sbjct 128 GYLSYTSKRDAQLFLKRVRKNLSKYSD--EKIRYYIVSEYGPKTFRAHYHVLFFYDEVKT 185
Query 163 AKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLF 222
K + + Q+W+ GRVD L+R + NSYVA Y+N +P + S +P S S F
Sbjct 186 QKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLP-RFLGDMSTKPFSCHSIRF 244
Query 223 GF-------EEVKKG 230
EE+ KG
Sbjct 245 ALGIHQSQKEEIYKG 259
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 103 bits (257), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 105/395 (27%), Positives = 167/395 (42%), Gaps = 51/395 (13%)
Query 103 GLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEI 162
G + Y+ D QLF KRLR Y++K+ EK+ + V EY P FRPH+H+L F SDE
Sbjct 115 GDVPYLRKTDLQLFLKRLRYYVTKQKPS-EKVRYFAVGEYGPVHFRPHYHLLLFLQSDEA 173
Query 163 AKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLF 222
+ + + ++W GRVD Q+++ Q ++YVA+Y+NS +IP V+KA S+ P F
Sbjct 174 LQICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKA-SSVCP-------F 225
Query 223 GFEEVKKGIQHASDKRSALFDGVP-------YISNQKFVRYVPSGSHIDRLFPRFTHYDG 275
K G +R ++ P + N K+ + S +PR +
Sbjct 226 SVHSQKLGQGFLDCQREKIYSLTPENFIRSSIVLNGKYKEFDVWRSCYSFFYPRCKGFVT 285
Query 276 SFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCEYNFRRGCQIKDFPDYMQ 335
R + Y + LF P K T E + ++ + + D Y
Sbjct 286 KSSRERAYSYSIYDTARLLF----PDAKTTFSLAKEIAIYIYYFHNPKETYLLDLYGYCS 341
Query 336 EFLHIVRLDGYSFLNWDVPI-----GKISRFFYR----------FNRFEAMKGSL---RS 377
+ + L Y F + DV + G+ SR+ +R F F +L +S
Sbjct 342 DQSKLYELSQY-FYDSDVLLHSFNSGEFSRYVHRIYTELLISKHFLYFVCTHNTLAERKS 400
Query 378 KLKAVSLFYDYRDYQSLKNQLSLQELVFAE--LGYSDELFDSFYVKPHIKVLKNAYID-- 433
K + + FY DY L Q+L + +G D D++ + N Y D
Sbjct 401 KQRLIEEFYSRLDYMHLTKFFEAQQLFYESDLIGDDDLCTDNWDNSYYPYFYNNVYTDTN 460
Query 434 --------KWKDVNYKEVHYFRVKHKVLNDENNIF 460
+ + K++ R+KHK LND N +F
Sbjct 461 LFEKTPVYRLYSSDVKKLFNDRIKHKKLNDANKVF 495
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 99.0 bits (245), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 104/395 (26%), Positives = 171/395 (43%), Gaps = 49/395 (12%)
Query 103 GLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEI 162
G L Y+ D QLF KR R Y++K+ K EK+ + + EY P FRPH+HIL F SDE
Sbjct 39 GYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQSDEA 97
Query 163 AKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLF 222
+ + V ++W GRVD QL++ + +SYVA Y+NS V +P V ++ P F
Sbjct 98 LQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVPKVLTL-PTLCP-------F 149
Query 223 GFEEVKKGIQHASDKRSALFDGVP-------YISNQKFVRYVPSGSHIDRLFPRFTHYDG 275
K G +R+ ++ P + N ++ + S FP+ +
Sbjct 150 CVHSQKLGQGFLQSERAKVYSLTPEQFVKRSIVINGRYKEFDVWRSAYAYFFPKCKGFAD 209
Query 276 SFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCEYNFRRGCQIKDFPDYMQ 335
R + Y + RLF P + T E V + ++ ++ D +
Sbjct 210 KSSRERAYSYGLYDTARRLF----PSAETTFALAKEIVGYIYYFHNKKDTYCLDIFGEVS 265
Query 336 EFLHIVRLDGYSF----LNWDVPIGKISRFFYR----------FNRFEAMKGSL---RSK 378
+ + + Y F +N+ + ++ R+ +R F F + +L + K
Sbjct 266 DQSDLYQFSQYFFEPEIVNYSLDSIEMCRYVHRVYTELLLSKHFLYFVCDRPTLSEQKRK 325
Query 379 LKAVSLFYDYRDYQSLKNQLSLQELVFAE--LGYSDELFDS--------FYVKPHI--KV 426
LK + FY DY LK Q+L + +G D + D+ FY + +V
Sbjct 326 LKLIEEFYSRLDYMHLKTFFENQQLFYESDLVGDLDLMSDAWENSYYPFFYDNVYFSSEV 385
Query 427 LKNAYIDKWKDVNYKEVHYFRVKHKVLNDENNIFL 461
K + + D+ ++ R+KHK LND N IF+
Sbjct 386 YKKTPVYRLYDMQISKLFSDRIKHKKLNDLNKIFV 420
>gi|575094340|emb|CDL65724.1| unnamed protein product [uncultured bacterium]
Length=486
Score = 81.3 bits (199), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 50/122 (41%), Positives = 73/122 (60%), Gaps = 12/122 (10%)
Query 111 RDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIA-KNFRQA 169
+D+ F KRLR L++ KI + SEY P T RPHFH +F+FDS ++ +FR A
Sbjct 157 KDFVNFVKRLRINLTRNYNYEGKITYFKCSEYGPTTNRPHFHGIFWFDSRALSFDSFRSA 216
Query 170 VYQSWRLGRVDTQ-----LAREQANSYVANYLNSVVSIP--FVYKAKKSIRPRSRFSNLF 222
V +SW++ D Q +ARE A +YVA+Y+N + S+P F++K +RP+ S F
Sbjct 217 VVESWKMCDKDKQYENVEIAREPA-TYVASYVNCLTSVPPLFLFKG---LRPKHSHSKGF 272
Query 223 GF 224
GF
Sbjct 273 GF 274
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 78.2 bits (191), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 59/184 (32%), Positives = 82/184 (45%), Gaps = 41/184 (22%)
Query 78 MLPEIAESLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSY 137
M P++ +K+ N + +KG Y++ R+ QLF KRLRKYL K G +KI +
Sbjct 96 MTPQLMNEYQKRVNYRIN-----YKGRFPYLSKRELQLFMKRLRKYLDKYEG--QKIRFF 148
Query 138 VVSEYSPKTFRPHFHILFFFDS-----------------------------DEIAKNFRQ 168
EY P +FRPHFHIL F D +
Sbjct 149 ATGEYGPLSFRPHFHILLFVDDPSLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLSKLEY 208
Query 169 AVYQSWRLGRVDTQ-LAREQANSYVANYLNSVVSIPFVYK--AKKSIRPRSRF--SNLFG 223
+ +SW G +D Q + + +SYVA Y+NS V +P K A KS SRF +FG
Sbjct 209 YIRESWPFGGIDAQSVEQGSCSSYVAGYVNSSVPLPSCLKVDAVKSFSQHSRFLGRKIFG 268
Query 224 FEEV 227
E +
Sbjct 269 TELI 272
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 77.4 bits (189), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 84/319 (26%), Positives = 133/319 (42%), Gaps = 61/319 (19%)
Query 41 LRYPNFISKFRPFILRSIPRVSKLQNFKDEYFEELVWMLPEIAESLKKKNNTDASGAFPQ 100
LR +FIS F S L NF +++ +++ + + K + + G
Sbjct 90 LRNDSFISDF----------CSDLHNFDNDFVDKMDYYSDYVINYESKYHKSCVYG---- 135
Query 101 FKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSD 160
GL + RD QLF KRLRK++ K G EKI Y++ EY K+ RPH+H L FF+S
Sbjct 136 -HGLYALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEYGTKSLRPHWHCLLFFNSS 192
Query 161 EIAKNFRQAVYQS---------------WRLGRVDTQLAREQANSYVANYLNSVVSIPFV 205
+++ F V W+ G D++ +A +YV++Y+N + P +
Sbjct 193 SLSQAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGEAYNYVSSYVNQSANFPKL 252
Query 206 Y------KAKKSIRPRSRFSNLFGFEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPS 259
KA SI+ S ++KG + +R D ++ R
Sbjct 253 LVLLSNQKAYHSIQLGQILSEQSIVSAIQKG-DFSFFERQFYLDTFGAANSYSVWR---- 307
Query 260 GSHIDRLFPRFTHYDGSFLRRSSQI-YEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCE 318
S+ R FP+FT SSQ+ YE RVL + E + + +C
Sbjct 308 -SYYSRFFPKFTC--------SSQLTYEQTYRVLTCY---ETLRDLFDTDSVGVICRRLF 355
Query 319 YNFRRGCQIKDFPDYMQEF 337
Y++ G +PDY F
Sbjct 356 YHYHFG-----YPDYHDIF 369
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 75.9 bits (185), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/138 (30%), Positives = 66/138 (48%), Gaps = 11/138 (8%)
Query 111 RDYQLFAKRLRKYLSKKI---GKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFR 167
+D Q F KRLR + K+ G +I ++ SEY P TFRPH+H + ++DS+ +
Sbjct 125 KDVQDFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEILHNELN 184
Query 168 QAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLFGFEEV 227
+ ++W+ G D L A+ YVA Y+N +P R+ F++ F
Sbjct 185 VLIRETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFL--------RTEFTSTFHLASK 236
Query 228 KKGIQHASDKRSALFDGV 245
I + D AL++ V
Sbjct 237 HPCIGYGKDDEEALYENV 254
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 72.0 bits (175), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 62/236 (26%), Positives = 102/236 (43%), Gaps = 10/236 (4%)
Query 74 ELVWMLPEIAESLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKY-- 131
E+VW + + N D + Y D F KRLR LS K+
Sbjct 81 EMVWTSNRLCDEKVIVGNYDFIKVSNSDVQAVAYCCKSDIVKFFKRLRSKLSYYFKKHHI 140
Query 132 ---EKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQA 188
EKI +V SEY PKT RPH+H + +FDS+E+A+ + + SW G D + A
Sbjct 141 ITNEKIRYFVCSEYGPKTLRPHYHAIIWFDSEEVARVIEKMLSSSWSNGFTDFEYVNSTA 200
Query 189 NSYVANYL--NSVVSIPFVYKAKKSIRPRSRFSNL-FGFEEVKKGIQHASDKRSALFDGV 245
YVA Y+ NSV+ + A ++ +S+ ++ + ++ +K + D F+
Sbjct 201 PQYVAKYVSGNSVLPEILQHDACRTFHLQSQAPSVGYRSDDYEKFEKEVIDGCYGHFEYD 260
Query 246 PYISNQKFVRYVPSGSHIDRLFPRFTHYDGSFLRRSSQIYEVVQRVLRLFARNEPF 301
+ FV+ P G+ R FP+ Y +IY + + ++ + P
Sbjct 261 SSSQSSVFVQ--PPGTLETRCFPKCREYRSLSRIEKLRIYAYKRDICSIYGIDTPI 314
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 71.2 bits (173), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/135 (30%), Positives = 71/135 (53%), Gaps = 7/135 (5%)
Query 76 VWMLPEIAESLKKKNNTDASGAFPQ---FKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYE 132
VW ++ES K +++ PQ + Y +D Q + KRLR + ++ K +
Sbjct 84 VWFSNRLSESGKFLSDSVCRSLPPQKMEDEVCFAYPCKKDVQDWFKRLRSAVDYQLNKNK 143
Query 133 ----KIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQA 188
+I ++ SEY P+TFRPH+H + ++DS+E+ +N + + ++W+ G L A
Sbjct 144 SNEFRIRYFICSEYGPRTFRPHYHAILWYDSEELQRNIGRLIRETWKNGNSVFSLVNNSA 203
Query 189 NSYVANYLNSVVSIP 203
+ YVA Y+N +P
Sbjct 204 SQYVAKYVNGDTRLP 218
>gi|546189465|ref|WP_021825245.1| hypothetical protein [Prevotella salivae]
gi|544001993|gb|ERK01417.1| hypothetical protein HMPREF9145_2741 [Prevotella salivae F0493]
Length=586
Score = 65.1 bits (157), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 67/123 (54%), Gaps = 8/123 (7%)
Query 96 GAFPQFKGLLKYVNIRDYQLFAKRLRKYLS----KKIGKYEKIHSYVVSEYSPKTFRPHF 151
G+ P FK L ++ Y L+ + YL+ KK + + ++ SEY+P TFRPHF
Sbjct 177 GSIP-FKEWLDDLDTETYDLYYSVYQYYLTDYEKKKESCKQSVRYFICSEYTPTTFRPHF 235
Query 152 HILFFFDSDEIAKNFRQAVYQSWRLG---RVDTQLAREQANSYVANYLNSVVSIPFVYKA 208
H LF+FD ++ + ++++W++ ++ Q A++YV+ Y+ ++P V +A
Sbjct 236 HGLFWFDDEKAFSYAPRCIFKAWKMCAEININVQPVSGDASAYVSKYVTGNSNLPPVLQA 295
Query 209 KKS 211
K +
Sbjct 296 KST 298
Lambda K H a alpha
0.325 0.140 0.424 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3125101418307