bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_5
Length=406
Score E
Sequences producing significant alignments: (Bits) Value
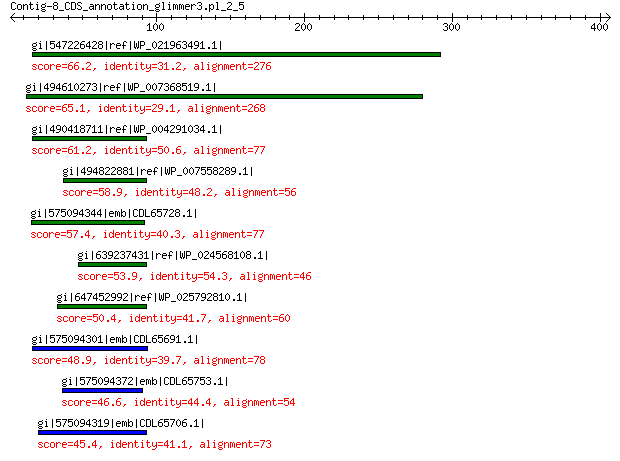
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 66.2 1e-08
gi|494610273|ref|WP_007368519.1| hypothetical protein 65.1 3e-08
gi|490418711|ref|WP_004291034.1| hypothetical protein 61.2 5e-07
gi|494822881|ref|WP_007558289.1| hypothetical protein 58.9 2e-06
gi|575094344|emb|CDL65728.1| unnamed protein product 57.4 8e-06
gi|639237431|ref|WP_024568108.1| hypothetical protein 53.9 6e-05
gi|647452992|ref|WP_025792810.1| hypothetical protein 50.4 0.001
gi|575094301|emb|CDL65691.1| unnamed protein product 48.9 0.005
gi|575094372|emb|CDL65753.1| unnamed protein product 46.6 0.023
gi|575094319|emb|CDL65706.1| unnamed protein product 45.4 0.054
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 66.2 bits (160), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 86/334 (26%), Positives = 134/334 (40%), Gaps = 64/334 (19%)
Query 16 AIFGLIGSVFGSSMSARSQRKANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKE----- 70
A+ G S G+ ++SQ N+ N +I QMNNE+N + K + DM+N++
Sbjct 8 ALIGAGASFLGNIFGSKSQSDTNKTNLQIAQMNNEYNERMFNKQLEYNQDMFNQQVEYDQ 67
Query 71 ---------------------NAYNTPSAQRERMEQAGYNAYM-------------Npad 96
YN+ AQR R+E AG N Y+ + +
Sbjct 68 KKMEQQNNFNARMQNEAIGAQQVYNSAKAQRARLEAAGLNPYLMMSGGNAGAVSAVSGSS 127
Query 97 agsasgmssttsasasspaVMQA--TDFSSLGEV-----------GVRLAQELKLFSEKK 143
S + +S AVMQA DFS + + GVR AQ L +
Sbjct 128 GSGGSPSPMGVNPPTASSAVMQAFRPDFSGVTGIIQTLLDIQAQKGVRDAQAFSLGEQAS 187
Query 144 GLDIRNFSLKDYLKAQIDKMRGETNWRNLSPEAI------RFNIMSGLEAAKIQMEGLKE 197
G I N + L I + + N +N S E++ R M + +K Q E
Sbjct 188 GFKIENKYKAEKLLWDIYNSKADYNLKN-SQESLNNMSFARLQAMFSSDVSKAQREAE-- 244
Query 198 QWINQKWSNNLLRANVANSLLDAEGKTVINKYLDQQQQADLNVKAAHYEELLLRGQLHSR 257
N +++ L+RA A L KY DQ+ +L + +A L+ G+
Sbjct 245 ---NAQFTGELIRAQTACQQLQGLLGAKELKYYDQKVLQELAIMSAQQYSLVAAGKASEA 301
Query 258 EARESLSRELLNYTRANGQKISNKVAERTANNLI 291
+AR+++ L + G K+ N V ++TAN LI
Sbjct 302 QARQAIENALNLVEQREGIKVDNYVKQKTANALI 335
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 65.1 bits (157), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 78/289 (27%), Positives = 133/289 (46%), Gaps = 39/289 (13%)
Query 12 PYDLAIFG--------LIGSVFGSSMSARSQRKANEMNFKINQMNNEFNAKEAEKARAFQ 63
P D +I G L+G +FG + +Q +ANE N +I + N+ + ++ AF
Sbjct 63 PIDPSIIGSAISGGVSLLGGLFGGHSNKTAQDRANETNLQIAREANQNQYQMFQEQNAFN 122
Query 64 LDMWNKENAYNTPSAQRERMEQAGYNAYM---NpadagsasgmssttsasasspaVMQAT 120
MWN+ N YN+P+AQ +R AG N Y+ N + S + S + VM AT
Sbjct 123 ERMWNQMNQYNSPAAQMQRYTDAGINPYIAAGNVQTGNAQSALQSAPAPQQHVAQVMPAT 182
Query 121 --------DFSSLGEVGVRLAQELKLFSEKKGLDIRNFSLKDYLKAQIDKMRGET-NWRN 171
F+ +G V + AQ ++ K D + AQ+ K+ ET N N
Sbjct 183 GMGDAVQNSFAQIGNVISQFAQNQLALAQAKKTDAEASWIDRLNSAQMGKLGAETLNIHN 242
Query 172 LSPEAIRFNIMSGLEAAKIQMEGLKEQWINQKWSNNLLRANVANSLLDAEGKTVINKYLD 231
N + GL+ +I+ + L + S + +A + N+L+DA+ + + +
Sbjct 243 Q-------NSLLGLD-YQIKSDTLGNYKLLSDLS--VQQAALTNNLVDAQTRKAL--FES 290
Query 232 QQQQADLNVKAAHYEELLLRGQLHSREARESLSRELLNYTRA-NGQKIS 279
+ ++KA + E+ +L +E ES+SR+ LNY A N +K++
Sbjct 291 DLAMVESHIKAKYGEKQVL------QEISESVSRQYLNYVSAHNSEKLT 333
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 61.2 bits (147), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 52/114 (46%), Gaps = 37/114 (32%)
Query 16 AIFGLIGSVFGSSMS----ARSQRKANEMNFKINQMNNEFNAKEAEKA------------ 59
A+ G++GS G+ S A + N+ N +I QMNN FN K +K
Sbjct 5 AMTGIVGSAIGAGTSLIGGASTTHMQNQANKEIAQMNNAFNEKMFDKQIAYNKEMYQTQL 64
Query 60 ---------------------RAFQLDMWNKENAYNTPSAQRERMEQAGYNAYM 92
+A+Q +MWNK+N YN PSAQR R+E AG N YM
Sbjct 65 GDQWKFYDDQKANAWKLYEDNKAYQTEMWNKQNEYNDPSAQRARLEAAGLNPYM 118
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 58.9 bits (141), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 37 ANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPSAQRERMEQAGYNAYM 92
N+ N +I QM+NE+N ++ E+ + DMWN EN YN+ S+QR+R+E+AG N YM
Sbjct 43 TNQANIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYM 98
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 57.4 bits (137), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 43/77 (56%), Gaps = 11/77 (14%)
Query 15 LAIFGLIGSVFGSSMSARSQRKANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKENAYN 74
L I + +FG S R F + QM E+ ++EA+K R FQLDMWN+ N YN
Sbjct 27 LGIVDGVAGMFGQSSDQR---------FALGQM--EWQSQEAQKQRDFQLDMWNRNNEYN 75
Query 75 TPSAQRERMEQAGYNAY 91
P Q +R+E+AG N +
Sbjct 76 KPDEQMKRLEEAGINPW 92
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 53.9 bits (128), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/46 (54%), Positives = 32/46 (70%), Gaps = 0/46 (0%)
Query 47 MNNEFNAKEAEKARAFQLDMWNKENAYNTPSAQRERMEQAGYNAYM 92
MNN N K A + RAF LDMWN+ N YNTP AQ +R+++AG N +
Sbjct 18 MNNSSNKKIARENRAFALDMWNRNNEYNTPLAQMQRLKEAGLNPNL 63
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 50.4 bits (119), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/60 (42%), Positives = 39/60 (65%), Gaps = 0/60 (0%)
Query 33 SQRKANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPSAQRERMEQAGYNAYM 92
S RK N+ N +I + N+ N + +++ AF M+++ NAYNTPSAQ +R +AG N Y+
Sbjct 25 SNRKTNQTNLQIARETNQMNYQLFQESNAFNEKMYHEANAYNTPSAQMQRYAEAGINPYI 84
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 48.9 bits (115), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 44/81 (54%), Gaps = 3/81 (4%)
Query 16 AIFGLIGSVFGSSMSARSQ-RKANEMNFKINQMNNEFNAKEAEKA--RAFQLDMWNKENA 72
AI G++G+ G S S +++ F+ +Q + N E A R F MW N
Sbjct 16 AISGVVGTATGIYDSVASLWGGSSQQKFERHQAEDARNFTHQENALQRDFARQMWKDTND 75
Query 73 YNTPSAQRERMEQAGYNAYMN 93
YNTP AQ++R+EQAG N Y+N
Sbjct 76 YNTPIAQKQRLEQAGMNPYVN 96
>gi|575094372|emb|CDL65753.1| unnamed protein product [uncultured bacterium]
Length=385
Score = 46.6 bits (109), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 34/54 (63%), Gaps = 0/54 (0%)
Query 36 KANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPSAQRERMEQAGYN 89
KA + N + + NEFNA EAEK RAFQ ++ + ++N+PS Q + M AG N
Sbjct 58 KARQFNSQQTALQNEFNASEAEKNRAFQKSLYERSLSWNSPSNQLKMMADAGLN 111
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 45.4 bits (106), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 30/80 (38%), Positives = 41/80 (51%), Gaps = 18/80 (23%)
Query 20 LIGSVF---GSSMSARSQ----RKANEMNFKINQMNNEFNAKEAEKARAFQLDMWNKENA 72
LI +F GS +A+ Q R+ N+ N +I NN+FN + MWN +N
Sbjct 35 LINGLFSSNGSKQAAKYQLQAVRETNQANREIADQNNKFNER-----------MWNLQNE 83
Query 73 YNTPSAQRERMEQAGYNAYM 92
YN P QR R+E AG N Y+
Sbjct 84 YNRPDMQRARLEAAGLNPYL 103
Lambda K H a alpha
0.313 0.127 0.360 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2614174527744