bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_2
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
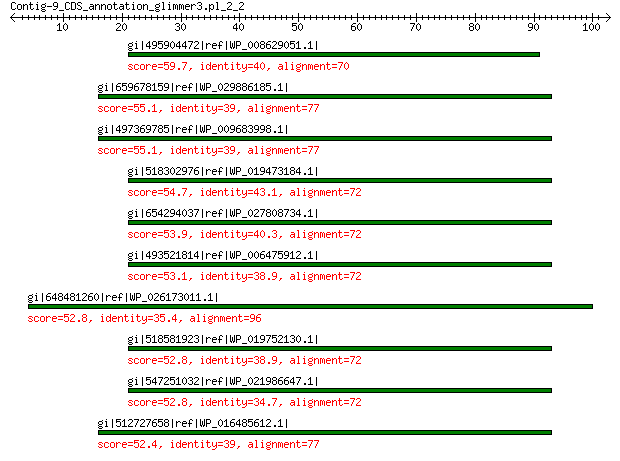
gi|495904472|ref|WP_008629051.1| hypothetical protein 59.7 3e-09
gi|659678159|ref|WP_029886185.1| peptidase M15 55.1 2e-07
gi|497369785|ref|WP_009683998.1| peptidase M15 55.1 2e-07
gi|518302976|ref|WP_019473184.1| MULTISPECIES: peptidase M15 54.7 2e-07
gi|654294037|ref|WP_027808734.1| peptidase M15 53.9 4e-07
gi|493521814|ref|WP_006475912.1| hypothetical protein 53.1 5e-07
gi|648481260|ref|WP_026173011.1| peptidase M15 52.8 1e-06
gi|518581923|ref|WP_019752130.1| peptidase M15 52.8 1e-06
gi|547251032|ref|WP_021986647.1| uncharacterized protein 52.8 1e-06
gi|512727658|ref|WP_016485612.1| peptidase M15 52.4 2e-06
>gi|495904472|ref|WP_008629051.1| hypothetical protein [Mariniradius saccharolyticus]
gi|472838632|gb|EMS32563.1| hypothetical protein C943_01290 [Mariniradius saccharolyticus
AK6]
Length=127
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 46/73 (63%), Gaps = 3/73 (4%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLIK-YRS 79
V S +R + N G++TSQHLTG A D++ + ++++ IK+S FDQLI+ +
Sbjct 38 VNSGYRAPRTNEAVGGSRTSQHLTGQAADIEVGHLTIEQLYQRIKNSGLPFDQLIQEFDQ 97
Query 80 FIHISF--ARGRK 90
++H+S+ A GRK
Sbjct 98 WVHVSYTAAGGRK 110
>gi|659678159|ref|WP_029886185.1| peptidase M15 [Pseudomonas putida]
Length=143
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 43/78 (55%), Gaps = 2/78 (3%)
Query 16 NIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLI 75
+ V+S +R+EK N GA SQH+ G A D + S E + I +S FDQLI
Sbjct 50 GVPIIVSSGYRSEKVNRLIGGAANSQHVQGLAADFTVIEVSPRETVQRISESTVPFDQLI 109
Query 76 -KYRSFIHISFARGRKPR 92
++ ++H+S ARG PR
Sbjct 110 LEFDKWVHLSVARG-TPR 126
>gi|497369785|ref|WP_009683998.1| peptidase M15 [Pseudomonas sp. TJI-51]
gi|324100352|gb|EGB98133.1| peptidase M15A [Pseudomonas sp. TJI-51]
Length=143
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 43/78 (55%), Gaps = 2/78 (3%)
Query 16 NIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLI 75
+ V+S +R+EK N GA SQH+ G A D + S E + I +S FDQLI
Sbjct 50 GVPIIVSSGYRSEKVNRLIGGAANSQHVQGLAADFTVIEVSPRETVQRISESTVPFDQLI 109
Query 76 -KYRSFIHISFARGRKPR 92
++ ++H+S ARG PR
Sbjct 110 LEFDKWVHLSVARG-TPR 126
>gi|518302976|ref|WP_019473184.1| MULTISPECIES: peptidase M15 [Pseudomonas]
Length=143
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 42/73 (58%), Gaps = 2/73 (3%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLI-KYRS 79
V+S FR+EK N GA SQH+ G A D + S E + I +S FDQLI ++
Sbjct 55 VSSGFRSEKVNRLIGGATNSQHVQGLAADFTVVEVSPRETVRRISESAVPFDQLILEFDR 114
Query 80 FIHISFARGRKPR 92
++H+S ARG PR
Sbjct 115 WVHLSVARG-TPR 126
>gi|654294037|ref|WP_027808734.1| peptidase M15 [Burkholderia cenocepacia]
Length=149
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 40/74 (54%), Gaps = 2/74 (3%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAID-LKPLYFSVDEFISMIKDSPFKFDQLIKYRS 79
+TS +R N G TS HL+G A D + P + S + I SP +FDQLI+ +
Sbjct 57 ITSGYRAAALNRAVGGVPTSAHLSGLAADFVCPKFGSPLDICRAISASPIEFDQLIQEGT 116
Query 80 FIHISFAR-GRKPR 92
++HI A G KPR
Sbjct 117 WVHIGLAPIGTKPR 130
>gi|493521814|ref|WP_006475912.1| hypothetical protein [Burkholderia cenocepacia]
gi|124870109|gb|EAY61825.1| hypothetical protein BCPG_00029 [Burkholderia cenocepacia PC184]
Length=104
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/74 (38%), Positives = 40/74 (54%), Gaps = 2/74 (3%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAID-LKPLYFSVDEFISMIKDSPFKFDQLIKYRS 79
+TS +R N G TS HL+G A D + P + + + I SP +FDQLI+ +
Sbjct 12 ITSGYRAAALNRAVGGVPTSAHLSGLAADFVCPKFGAPLDICRAISASPIEFDQLIQEGT 71
Query 80 FIHISFAR-GRKPR 92
++HI A G KPR
Sbjct 72 WVHIGLAPIGTKPR 85
>gi|648481260|ref|WP_026173011.1| peptidase M15 [Asticcacaulis benevestitus]
Length=151
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 49/97 (51%), Gaps = 3/97 (3%)
Query 4 ELMKFVEWLLHRNIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAID-LKPLYFSVDEFIS 62
+ M V LL + I V S +R+ N GA TS H+ G A+D + P Y + +
Sbjct 44 QQMDRVRHLLGKPIR--VNSGYRSPALNAAVGGAPTSAHMAGYAVDFVCPGYGTPLQICQ 101
Query 63 MIKDSPFKFDQLIKYRSFIHISFARGRKPRLMELNFT 99
I S ++DQLI+ S++HISF + R M FT
Sbjct 102 KIVASDIRYDQLIQEGSWVHISFDPRLRMRQMTATFT 138
>gi|518581923|ref|WP_019752130.1| peptidase M15 [Pseudomonas putida]
Length=143
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 42/73 (58%), Gaps = 2/73 (3%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLI-KYRS 79
++S +R+E+ N GA SQH+ G A D + FS E + I +S FDQLI ++
Sbjct 55 ISSGYRSERVNRLIGGASDSQHVQGLAADFTVIEFSPRETVRRISESALPFDQLILEFDK 114
Query 80 FIHISFARGRKPR 92
++H+S RG PR
Sbjct 115 WVHLSVTRG-TPR 126
>gi|547251032|ref|WP_021986647.1| uncharacterized protein [Odoribacter sp. CAG:788]
gi|524136061|emb|CCZ05909.1| uncharacterized protein BN783_00193 [Odoribacter sp. CAG:788]
Length=150
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 39/80 (49%), Gaps = 8/80 (10%)
Query 21 VTSAFRTEKQNNECNGAKTSQHLTGDAIDL--------KPLYFSVDEFISMIKDSPFKFD 72
+ S +R+ N G TSQH+ G+A D+ K +Y + E + ++ S FD
Sbjct 57 INSGYRSPALNGLVGGVATSQHVRGEAADVVFFRKEGGKTIYIASVEVLRVLTASGLNFD 116
Query 73 QLIKYRSFIHISFARGRKPR 92
Q I Y F+H+S+ R R
Sbjct 117 QAIAYNGFVHLSYTTARPNR 136
>gi|512727658|ref|WP_016485612.1| peptidase M15 [Pseudomonas putida]
gi|512577385|ref|YP_008093364.1| peptidase M15 [Pseudomonas putida H8234]
gi|511520885|gb|AGN78426.1| peptidase M15 [Pseudomonas putida H8234]
gi|551054298|gb|ERT19971.1| peptidase M15 [Pseudomonas putida SJ3]
Length=143
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 41/78 (53%), Gaps = 2/78 (3%)
Query 16 NIHFTVTSAFRTEKQNNECNGAKTSQHLTGDAIDLKPLYFSVDEFISMIKDSPFKFDQLI 75
+ V+S FR+EK N GA SQH+ G A D + S E I +S FDQLI
Sbjct 50 GVPIIVSSGFRSEKVNRLIGGATNSQHIQGLAADFTVMEVSPRETARRINESAVPFDQLI 109
Query 76 -KYRSFIHISFARGRKPR 92
++ ++H+S RG PR
Sbjct 110 LEFDRWVHLSVTRG-TPR 126
Lambda K H a alpha
0.325 0.138 0.415 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 436588657356