bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-14_CDS_annotation_glimmer3.pl_2_3
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
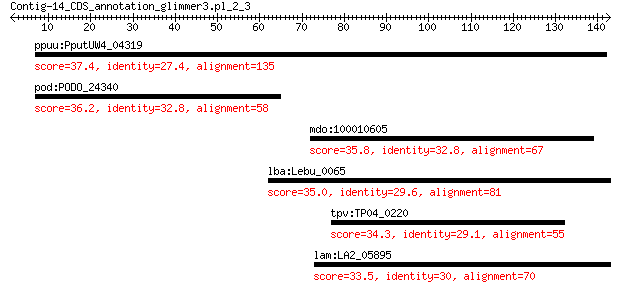
ppuu:PputUW4_04319 chemotaxis sensory transducer, Cache sensor 37.4 0.59
pod:PODO_24340 hypothetical protein 36.2 1.7
mdo:100010605 LAMB1; laminin, beta 1 35.8
lba:Lebu_0065 ethanolamine ammonia lyase large subunit 35.0 4.1
tpv:TP04_0220 hypothetical protein 34.3 6.6
lam:LA2_05895 phage-like protein 33.5 9.2
> ppuu:PputUW4_04319 chemotaxis sensory transducer, Cache sensor
Length=629
Score = 37.4 bits (85), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 62/135 (46%), Gaps = 3/135 (2%)
Query 7 INVLSNSEFTTVRPIDELRFVEELDGSIRFVSDATLLLNEQRIINDIGEDNYRNFIRQLQ 66
+ + +N EF T+ RFVE + GSIR VS AT +NE + + N F Q
Sbjct 325 LTIQNNDEFGTLGTAFN-RFVERIHGSIREVSSATGQVNEVA-LRVVAASNSSMFNSDQQ 382
Query 67 VNPSSPYKDAKFTDEQLMTEIKSRYVQSPSEVREWVRDMLDKQEQISDEVKAKLEEMKAS 126
+ +S A EI Q+ S+ + R + + +Q+ D + ++ +
Sbjct 383 ASRTSSVAAAINQLGAAAQEIARNAAQASSQASD-ARSLAEDGQQVVDRSIVAMNQLSSM 441
Query 127 ESESSESIETVESKS 141
S SS +IE++ SK+
Sbjct 442 LSASSTNIESLNSKT 456
> pod:PODO_24340 hypothetical protein
Length=617
Score = 36.2 bits (82), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 34/61 (56%), Gaps = 5/61 (8%)
Query 7 INVLSNSEFTTVRPIDELRFVEELDGSIRFVSDATLLLNEQRI---INDIGEDNYRNFIR 63
IN++ FT ++ ID++ + E G + V D L++ +I + DI DNY+ FI+
Sbjct 550 INIIITHRFTNIKNIDQIIVIHE--GKVDAVGDHDFLISNNKIYAELYDIQADNYKGFIK 607
Query 64 Q 64
+
Sbjct 608 E 608
> mdo:100010605 LAMB1; laminin, beta 1
Length=1786
Score = 35.8 bits (81), Expect = 2.7, Method: Composition-based stats.
Identities = 22/69 (32%), Positives = 43/69 (62%), Gaps = 2/69 (3%)
Query 72 PYKDAKFTDEQLMTEIKSRYVQSP-SEVREWVRDMLDKQEQISDEVKAKLEEMKASESE- 129
PY++ E+ + EIKS QSP +E + + ++ ++ E+++ +V K+ E++A S+
Sbjct 1215 PYRETVDNVEKKINEIKSILAQSPAAEPLKNIGNLFEEAEKLTKDVTEKMSEVEAKLSDA 1274
Query 130 SSESIETVE 138
+S+S TVE
Sbjct 1275 ASKSNSTVE 1283
> lba:Lebu_0065 ethanolamine ammonia lyase large subunit
Length=454
Score = 35.0 bits (79), Expect = 4.1, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 46/88 (52%), Gaps = 10/88 (11%)
Query 62 IRQLQVNPSSPYKDAKFT-------DEQLMTEIKSRYVQSPSEVREWVRDMLDKQEQISD 114
+R L+ NP+ PY++ + T +E++ EIK+ V SE+REW+ D + I
Sbjct 56 LRDLRNNPAVPYEEDEVTRIIQDAVNEKIYGEIKNWTV---SELREWILDSHTSGDDIKR 112
Query 115 EVKAKLEEMKASESESSESIETVESKSE 142
K EM A+ ++ +++ ++ S+
Sbjct 113 ISKGLTSEMVAAVAKLMSNMDLIQGASK 140
> tpv:TP04_0220 hypothetical protein
Length=434
Score = 34.3 bits (77), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 29/55 (53%), Gaps = 0/55 (0%)
Query 77 KFTDEQLMTEIKSRYVQSPSEVREWVRDMLDKQEQISDEVKAKLEEMKASESESS 131
KF +QL ++K+ +Q S+ EW+ D + K + E L E SE++++
Sbjct 132 KFNKQQLTNKLKTYLLQHASDFTEWLYDFVQKTVEYYHETHLPLNEENLSETQTT 186
> lam:LA2_05895 phage-like protein
Length=181
Score = 33.5 bits (75), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 43/75 (57%), Gaps = 5/75 (7%)
Query 73 YKDAKFTD--EQLMTEIKSRYVQSP---SEVREWVRDMLDKQEQISDEVKAKLEEMKASE 127
Y D +F + + TE ++SP ++V WV D D Q +I + +AK+E + S+
Sbjct 47 YSDVEFAELPWHVHTEKPDENLKSPVWSNDVNGWVEDSKDGQAEILAQAQAKIEALDKSK 106
Query 128 SESSESIETVESKSE 142
+E ++++T++++ E
Sbjct 107 AELDQTVKTLQTQQE 121
Lambda K H a alpha
0.309 0.127 0.328 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125938487780