bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_4
Length=270
Score E
Sequences producing significant alignments: (Bits) Value
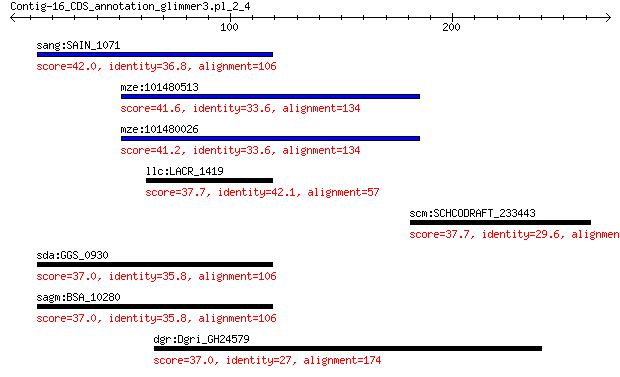
sang:SAIN_1071 hypothetical protein 42.0 0.083
mze:101480513 GTPase IMAP family member 7-like 41.6 0.11
mze:101480026 GTPase IMAP family member 7-like 41.2 0.14
llc:LACR_1419 hypothetical protein 37.7 1.8
scm:SCHCODRAFT_233443 hypothetical protein 37.7 2.6
sda:GGS_0930 hypothetical protein 37.0 3.1
sagm:BSA_10280 hypothetical protein 37.0 3.1
dgr:Dgri_GH24579 GH24579 gene product from transcript GH24579-RA 37.0 4.4
> sang:SAIN_1071 hypothetical protein
Length=288
Score = 42.0 bits (97), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 39/126 (31%), Positives = 58/126 (46%), Gaps = 31/126 (25%)
Query 13 FMTLTFSEEKLEEYKKLCNSEDPNTIATKAMRLMLERCRRKLKHSIKHWFITELGHNGTE 72
F TLTF + K++ + D A K +R L+ R K ++ F+ EL +G
Sbjct 73 FWTLTFDDNKVD-------ARD-YPYAKKRLRAWLKYQREKY-GKFQYIFVAELHKSG-- 121
Query 73 RMHLHGLVWGIG----------MDKLVEEK---------WQNGIVFTGTFVN-EKTINYI 112
R+H HGL G ++L+++K W+NG + EKT NYI
Sbjct 122 RIHFHGLTAGFSPPLTEARSPKTNRLIKKKGLQIYNAETWKNGFSTVSKIQDREKTANYI 181
Query 113 TKYITK 118
+KYITK
Sbjct 182 SKYITK 187
> mze:101480513 GTPase IMAP family member 7-like
Length=308
Score = 41.6 bits (96), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 45/138 (33%), Positives = 61/138 (44%), Gaps = 18/138 (13%)
Query 51 RRKLKHSIKHWF--ITELGHNGTERMHLHGLVWGIGMDKLVEEKWQNGIV-FT-GTFVNE 106
R KL+ H F + LG E H L+ K W IV FT G + +
Sbjct 97 RLKLQEPGPHVFAYVIPLGRLTQEDQDTHTLIEA----KFGPRVWDYTIVLFTHGDRLED 152
Query 107 KTINYITKYITKTDEYHKEFIGQILCSAGIGNGYTQRSDAQKHTYKKGETIETYRLRNGA 166
KTIN I IT++DE + FI + CS G + + QK E IET NG
Sbjct 153 KTINNI---ITESDENLRNFIRK--CSGGFHVFNNKTPEDQKQVTTFMEKIETLVALNGG 207
Query 167 KINLPTYYRNKLFTEEER 184
+YY+ +L+ E+ER
Sbjct 208 -----SYYKTELYPEKER 220
> mze:101480026 GTPase IMAP family member 7-like
Length=337
Score = 41.2 bits (95), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 45/138 (33%), Positives = 61/138 (44%), Gaps = 18/138 (13%)
Query 51 RRKLKHSIKHWF--ITELGHNGTERMHLHGLVWGIGMDKLVEEKWQNGIV-FT-GTFVNE 106
R KL+ H F + LG E H L+ K W IV FT G + +
Sbjct 97 RLKLQEPGPHVFAYVIPLGRLTQEDQDTHTLIEA----KFGPRVWDYTIVLFTHGDRLED 152
Query 107 KTINYITKYITKTDEYHKEFIGQILCSAGIGNGYTQRSDAQKHTYKKGETIETYRLRNGA 166
KTIN I IT++DE + FI + CS G + + QK E IET NG
Sbjct 153 KTINNI---ITESDENLRNFIRK--CSGGFHVFNNKTPEDQKQVTTFMEKIETLVALNGG 207
Query 167 KINLPTYYRNKLFTEEER 184
+YY+ +L+ E+ER
Sbjct 208 -----SYYKTELYPEKER 220
> llc:LACR_1419 hypothetical protein
Length=284
Score = 37.7 bits (86), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 34/77 (44%), Gaps = 21/77 (27%)
Query 62 FITELGHNGTERMHLHGLVWGI-------------------GMDKLVEEKWQNGIVFTGT 102
F+ EL H +ER+H HG+ G GM +W+NG
Sbjct 112 FVPEL-HKSSERIHFHGVTQGFSPPLVEARYPKNRRLIKRNGMQIYNAPRWKNGFSTVSR 170
Query 103 FVN-EKTINYITKYITK 118
+ EK+ +YITKYI+K
Sbjct 171 IQSKEKSASYITKYISK 187
> scm:SCHCODRAFT_233443 hypothetical protein
Length=1773
Score = 37.7 bits (86), Expect = 2.6, Method: Composition-based stats.
Identities = 24/81 (30%), Positives = 40/81 (49%), Gaps = 3/81 (4%)
Query 181 EEEREKLWIDKIEKGIVWVMGQKVKIDNTEEYEALLEQARHDAIRLQGHQEQNWDKQKYF 240
+EE+ + K EK + ++ K EE E+ R + R + QEQ ++ K
Sbjct 1616 KEEKAR---QKAEKEVAKAAEKEGKARAKEEQAREKERVRQEKARAKEDQEQEKERAKRE 1672
Query 241 KRLEKQRKKQQKELRKWENHM 261
K L KQR+ +QK+L K + +
Sbjct 1673 KELTKQREAEQKQLAKQQKEL 1693
> sda:GGS_0930 hypothetical protein
Length=272
Score = 37.0 bits (84), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 54/126 (43%), Gaps = 31/126 (25%)
Query 13 FMTLTFSEEKLEEYKKLCNSEDPNTIATKAMRLMLERCRRKLKHSIKHWFITELGHNGTE 72
F TLTF + K+ + L I K +R L+ R K ++ FI EL NG
Sbjct 74 FWTLTFDDAKINSHDYL--------IVKKKIRTWLKAQREKYG-KFRYIFIPELHKNG-- 122
Query 73 RMHLHGLVWGIG----------MDKLVE---------EKWQNGI-VFTGTFVNEKTINYI 112
R+H HG+ G +L++ + +Q G T ++K NYI
Sbjct 123 RLHFHGVTGGFSPKLTKARNTKTGRLIKKNGKQVYNVDSYQLGFSTVTKIDSSKKVANYI 182
Query 113 TKYITK 118
TKYITK
Sbjct 183 TKYITK 188
> sagm:BSA_10280 hypothetical protein
Length=272
Score = 37.0 bits (84), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 54/126 (43%), Gaps = 31/126 (25%)
Query 13 FMTLTFSEEKLEEYKKLCNSEDPNTIATKAMRLMLERCRRKLKHSIKHWFITELGHNGTE 72
F TLTF + K+ + L I K +R L+ R K ++ FI EL NG
Sbjct 74 FWTLTFDDAKINSHDYL--------IVKKKIRTWLKAQREKYG-KFRYIFIPELHKNG-- 122
Query 73 RMHLHGLVWGIG----------MDKLVE---------EKWQNGI-VFTGTFVNEKTINYI 112
R+H HG+ G +L++ + +Q G T ++K NYI
Sbjct 123 RLHFHGVTGGFSPKLTKARNTKTGRLIKKNGKQVYNVDSYQLGFSTVTKIDSSKKVANYI 182
Query 113 TKYITK 118
TKYITK
Sbjct 183 TKYITK 188
> dgr:Dgri_GH24579 GH24579 gene product from transcript GH24579-RA
Length=1030
Score = 37.0 bits (84), Expect = 4.4, Method: Composition-based stats.
Identities = 47/188 (25%), Positives = 78/188 (41%), Gaps = 39/188 (21%)
Query 66 LGHNGTERMHLHGLVW------GIGMDKLVEEKWQNGIVFT-------GTFV-NEKTINY 111
L H T R+ LHG W G +KL+ E+ +NG G FV + +T +
Sbjct 244 LAHLATMRVQLHGYTWFHGNLSGKEAEKLILERGKNGSFLVRESQSKPGDFVLSVRTEDK 303
Query 112 ITKYITKTDEYHKEFIGQILCSAGIGNGYTQRSDAQKHTYKKGETIETYRLRNGAKINLP 171
+T + + + + G G + S+ +H YK+ +ET G ++L
Sbjct 304 VTHVMIRWQDTKYD--------VGGGESFATLSELIEH-YKRHPMVETC----GTVVHLR 350
Query 172 TYYRNKLFTEEEREKLWIDKIEKGIVWVMGQKVKIDNTEEYEALLEQARHDAIRLQGHQE 231
+ I GI + Q VK EE+E+L + +R RL+G++E
Sbjct 351 QPFNATRI------------IAAGINARVEQLVKGGFWEEFESLQQDSRDTFSRLEGYKE 398
Query 232 QNWDKQKY 239
+N K +Y
Sbjct 399 ENKSKNRY 406
Lambda K H a alpha
0.317 0.133 0.405 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 421746798214