bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_7
Length=639
Score E
Sequences producing significant alignments: (Bits) Value
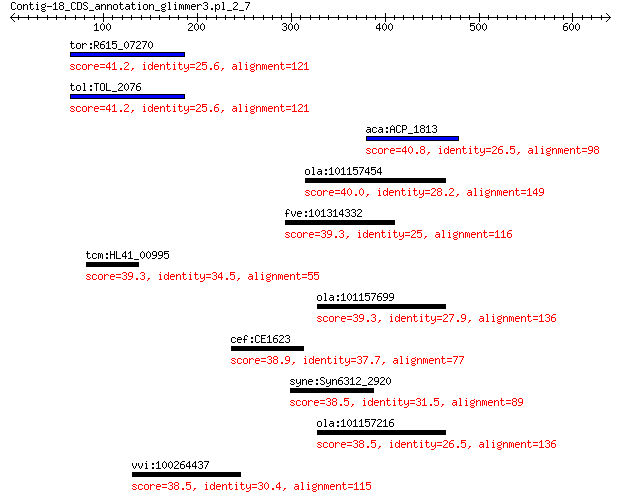
tor:R615_07270 Na(+)-translocating NADH-quinone reductase subu... 41.2 0.69
tol:TOL_2076 Na(+)-translocating NADH-quinone reductase subunit A 41.2 0.69
aca:ACP_1813 ImpB/MucB/SamB family protein 40.8 0.85
ola:101157454 uncharacterized LOC101157454 40.0 1.6
fve:101314332 capsid protein VP1-like 39.3 2.4
tcm:HL41_00995 hypothetical protein 39.3 3.1
ola:101157699 transmembrane protease serine 9-like 39.3 3.2
cef:CE1623 hypothetical protein 38.9 3.4
syne:Syn6312_2920 cobalamin biosynthesis protein CbiD 38.5 4.6
ola:101157216 transmembrane protease serine 9-like 38.5 5.6
vvi:100264437 methionine S-methyltransferase-like 38.5 6.1
> tor:R615_07270 Na(+)-translocating NADH-quinone reductase subunit
A
Length=444
Score = 41.2 bits (95), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 53/127 (42%), Gaps = 9/127 (7%)
Query 65 LMNGFKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIA 124
+ +G K+ +A PD Y M+ + + K G+V F Y PA +
Sbjct 20 ITDGPKVTQVAVMGPD---YVGMKPTMAVQEGDRVKKGQVLFTDKKTEGVQYTAPAAGVV 76
Query 125 RRVRR------LTFGLELNADRAKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPG 178
+ + R L+ +EL+ D + + S+ ++LNS ++ GLW L P
Sbjct 77 KAINRGARRVFLSVVIELDGDEEETFTSYTPEQLNSLDAKDVEANLVASGLWTALRTRPY 136
Query 179 AVCPNLG 185
+ P LG
Sbjct 137 SKVPALG 143
> tol:TOL_2076 Na(+)-translocating NADH-quinone reductase subunit
A
Length=444
Score = 41.2 bits (95), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 53/127 (42%), Gaps = 9/127 (7%)
Query 65 LMNGFKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIA 124
+ +G K+ +A PD Y M+ + + K G+V F Y PA +
Sbjct 20 ITDGPKVTQVAVMGPD---YVGMKPTMAVQEGDRVKKGQVLFTDKKTEGVQYTAPAAGVV 76
Query 125 RRVRR------LTFGLELNADRAKIYESWVSDELNSTGDDGQPTHIGRGGLWDWLGIAPG 178
+ + R L+ +EL+ D + + S+ ++LNS ++ GLW L P
Sbjct 77 KAINRGARRVFLSVVIELDGDEEETFTSYTPEQLNSLDAKDVEANLVASGLWTALRTRPY 136
Query 179 AVCPNLG 185
+ P LG
Sbjct 137 SKVPALG 143
> aca:ACP_1813 ImpB/MucB/SamB family protein
Length=441
Score = 40.8 bits (94), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 26/98 (27%), Positives = 45/98 (46%), Gaps = 6/98 (6%)
Query 380 DVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYC 439
+ R LW ++ + P KP F+GVW ++ P ++ ++ S+ +A L +DR
Sbjct 349 EALRKLWELRPTGPAHQKPFFVGVWLGNLVPDHLHTLSLFSSLETEARRTSLTTTIDR-- 406
Query 440 DFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDL 477
+ G+D A P + +L P F + PDL
Sbjct 407 -VNLKYGLDTLA--PASMLLAKAAAPTRIAFTNI-PDL 440
> ola:101157454 uncharacterized LOC101157454
Length=620
Score = 40.0 bits (92), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 42/157 (27%), Positives = 65/157 (41%), Gaps = 21/157 (13%)
Query 315 SVPYSPDLFSN-------IIKQGSSP-AVEIEVMNALDASTDTGFSVAVPELRLKTKLQN 366
S+P SP + S + + GS+P V + V N + S TG +VAV +L + L N
Sbjct 401 SLPKSPPVASEWTVVLGRLKQNGSNPFEVSLNVTN-ITLSNQTGSNVAVLQLSTRPPLNN 459
Query 367 WMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDA 426
++ + + GR V T W S+ K + L +Q S+ AN S
Sbjct 460 YIQPICLDNGRTFPVGTTCWAAGWSSGRGGKEEVLQEFQTSVLECPTSTAANPS------ 513
Query 427 NLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAML 463
C R+ G SG K+ G++ A+L
Sbjct 514 ------ICTGRFTLEQGDSGGPLMCKQDGSWHQAAVL 544
> fve:101314332 capsid protein VP1-like
Length=421
Score = 39.3 bits (90), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 55/120 (46%), Gaps = 5/120 (4%)
Query 294 IPINCVQAMACAGIQGYG-GLLSVPYSPDLFSNIIKQGSSPAVEI---EVMNALDASTDT 349
+P+ +A +G G G++S +L N+ GS +++I V L A
Sbjct 244 LPLGTSAPIAFSGASGSDVGVISTTQG-NLIKNMYSTGSGTSLKIGSATVATGLYADLSA 302
Query 350 GFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASIN 409
+ + +LR ++Q ++R G R ++ R+ +GV S + +P++LG IN
Sbjct 303 ATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPIN 362
> tcm:HL41_00995 hypothetical protein
Length=1074
Score = 39.3 bits (90), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 34/56 (61%), Gaps = 3/56 (5%)
Query 82 AIYGWMRNGRRYEPDEYTKFGKVYFPLAGP-NIGNYIDPAFQIARRVRRLTFGLEL 136
+ ++++GR + D++ GK+ F ++GP + GN+ DPA+Q RRV L +L
Sbjct 368 TVKAYIKDGRLFVEDKFRGPGKISFSISGPFDFGNFSDPAYQ--RRVEELNLAGKL 421
> ola:101157699 transmembrane protease serine 9-like
Length=692
Score = 39.3 bits (90), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 38/137 (28%), Positives = 57/137 (42%), Gaps = 14/137 (10%)
Query 328 KQGSSP-AVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLW 386
+ GS+P V + V N + S TG +VAV +L L N++ + + GR V T W
Sbjct 371 QNGSNPFEVSLNVTN-ITLSNQTGSNVAVLQLSTPPPLNNYIQPICLDKGRTFPVGTTCW 429
Query 387 GVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYCDFSGHSG 446
S+ K + L +Q S+ ANGS C R+ G SG
Sbjct 430 AAGWSSGRGGKEEVLQEFQTSVLECPTSTAANGS------------ICTGRFTLEQGDSG 477
Query 447 IDYYAKEPGTFMLIAML 463
K+ G++ A+L
Sbjct 478 GPLLCKQDGSWYQAAVL 494
> cef:CE1623 hypothetical protein
Length=489
Score = 38.9 bits (89), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 29/86 (34%), Positives = 43/86 (50%), Gaps = 9/86 (10%)
Query 236 GEMQRVRPDGQQESLYRPFFSDVFTS--FKPDDFLLYLDTLSYRTRVGAEIDALTLQLEG 293
GE++R+RPD + L+RP +S F P+ LYLD S+ AE D+LT +
Sbjct 35 GEVRRLRPDEIAKYLFRPVVDKYSSSNIFIPNASRLYLDVGSHPEYATAECDSLTQLINA 94
Query 294 -----IPIN--CVQAMACAGIQGYGG 312
+ +N V+A A +G GG
Sbjct 95 EKAGDVIVNRMAVEAEAALAREGIGG 120
> syne:Syn6312_2920 cobalamin biosynthesis protein CbiD
Length=373
Score = 38.5 bits (88), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 40/92 (43%), Gaps = 8/92 (9%)
Query 299 VQAMACAGIQGYGGLLSVPYSPDLFSNIIKQGSSPAVEIEVM---NALDASTDTGFSVAV 355
V ++ G G LS P D+F + Q + + ++ N LD +T G
Sbjct 162 VSGLSLLGTSGISQALSAPEQLDIFRQELLQKAQTSKQLVFCIGENGLDLATKAGIP--- 218
Query 356 PELRLKTKLQNWMDRLFVSGGRVGDVFRTLWG 387
PEL++KT NW+ L V G G LWG
Sbjct 219 PELQVKTA--NWLGPLLVEAGLQGVNSLVLWG 248
> ola:101157216 transmembrane protease serine 9-like
Length=600
Score = 38.5 bits (88), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 14/137 (10%)
Query 328 KQGSSP-AVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLW 386
+ GS+P V + V N + S TG +VAV +L + L N++ + + GR + T W
Sbjct 375 QNGSNPFEVSLNVTN-ITLSNQTGSNVAVLQLSTQPPLNNYIQPICLDNGRTFPLGTTCW 433
Query 387 GVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYCDFSGHSG 446
S+ + + L +Q S+ ANGS C R+ G SG
Sbjct 434 AAGWSSGRGGEEEVLQEFQTSVLECPTSTAANGS------------ICTGRFTLQQGDSG 481
Query 447 IDYYAKEPGTFMLIAML 463
K+ G++ A+L
Sbjct 482 GPLMCKQDGSWHQAAVL 498
> vvi:100264437 methionine S-methyltransferase-like
Length=1092
Score = 38.5 bits (88), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 35/126 (28%), Positives = 54/126 (43%), Gaps = 17/126 (13%)
Query 131 TFGLELNADRAKIYESWVSDELNSTGDDGQPTHIGRGG-LWDWLGIAPG---AVCPNLGI 186
+GL++N KI SW++ LN+ D+GQP + G L D + A C + GI
Sbjct 145 VYGLDINPRAVKI--SWINLYLNALDDNGQPIYDGENKTLLDRVEFHESDLLAYCRDRGI 202
Query 187 KLPSVAGVRGQFYPPS-------FKFNGAPFFAYFLSHYYYIANMQEKYMYFTRGVGEMQ 239
+L + G Q P+ N + F Y LS+Y + E G+G +
Sbjct 203 ELERIVGCIPQILNPNPDAMSKMITENASEEFLYSLSNYCALQGFVEDQF----GLGLIA 258
Query 240 RVRPDG 245
R +G
Sbjct 259 RAVEEG 264
Lambda K H a alpha
0.321 0.140 0.442 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1500132135889