bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_8
Length=222
Score E
Sequences producing significant alignments: (Bits) Value
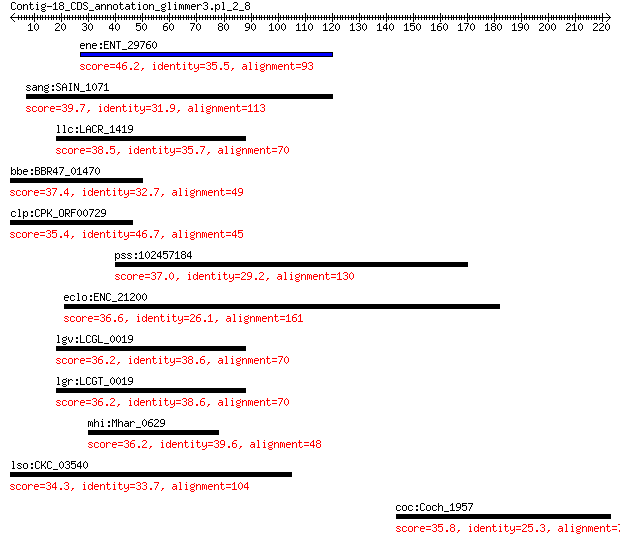
ene:ENT_29760 hypothetical protein 46.2 0.002
sang:SAIN_1071 hypothetical protein 39.7 0.33
llc:LACR_1419 hypothetical protein 38.5 0.79
bbe:BBR47_01470 hypothetical protein 37.4 1.4
clp:CPK_ORF00729 hypothetical protein 35.4 2.6
pss:102457184 XRCC3; X-ray repair complementing defective repa... 37.0 2.7
eclo:ENC_21200 hypothetical protein 36.6 3.2
lgv:LCGL_0019 hypothetical protein 36.2 3.9
lgr:LCGT_0019 hypothetical protein 36.2 3.9
mhi:Mhar_0629 adenine deaminase 36.2 5.9
lso:CKC_03540 hypothetical protein 34.3 6.2
coc:Coch_1957 coproporphyrinogen III oxidase (EC:1.3.99.22) 35.8 7.2
> ene:ENT_29760 hypothetical protein
Length=299
Score = 46.2 bits (108), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 46/106 (43%), Gaps = 15/106 (14%)
Query 27 GRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASR--------WKYGFVFVGYVS--EETC 76
G H+H +L D+P I+ P PLL + W YGF + E T
Sbjct 136 GAVHFHSLLGDIPANFIEAKHPKTGA--PLLRNERQIYNLADWTYGFTDCEEIEDKERTA 193
Query 77 SYITKYVTKSINGDKV---RPRIISSFGIGSNYLDTEEAVLHKLGN 119
SY+TKY+TK + DK + R +S G+ +D A L N
Sbjct 194 SYLTKYITKELMTDKEMFRKKRYFNSKGLKKPQIDYINAETDNLKN 239
> sang:SAIN_1071 hypothetical protein
Length=288
Score = 39.7 bits (91), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 54/124 (44%), Gaps = 13/124 (10%)
Query 7 RKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDV------PGHHPLLASR 60
R+ Y K ++ FV E GR H+HG+ L + SP G A
Sbjct 103 REKYGK-FQYIFVAELHKS-GRIHFHGLTAGFSPPLTEARSPKTNRLIKKKGLQIYNAET 160
Query 61 WKYGFVFVGYVS--EETCSYITKYVTK---SINGDKVRPRIISSFGIGSNYLDTEEAVLH 115
WK GF V + E+T +YI+KY+TK +I +PR S G+ ++ +
Sbjct 161 WKNGFSTVSKIQDREKTANYISKYITKELMAIPSGYHQPRYFVSRGLNQPDIEYKTIADK 220
Query 116 KLGN 119
+L N
Sbjct 221 ELAN 224
> llc:LACR_1419 hypothetical protein
Length=284
Score = 38.5 bits (88), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 35/78 (45%), Gaps = 8/78 (10%)
Query 18 FVCEFGTLYGRPHYHGILFDVPQTLIDGYSPD------VPGHHPLLASRWKYGFVFVGYV 71
FV E R H+HG+ L++ P G A RWK GF V +
Sbjct 112 FVPELHKSSERIHFHGVTQGFSPPLVEARYPKNRRLIKRNGMQIYNAPRWKNGFSTVSRI 171
Query 72 S--EETCSYITKYVTKSI 87
E++ SYITKY++K +
Sbjct 172 QSKEKSASYITKYISKDL 189
> bbe:BBR47_01470 hypothetical protein
Length=254
Score = 37.4 bits (85), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query 1 LFLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFD-VPQTLIDGYSPD 49
L L R+D+ + + W++ E+G+ Y +YH ILF V + ++ SP+
Sbjct 60 LQLSNCRRDFDRAVEEWYITEYGSGYKGVNYHDILFTLVKEAVVQYQSPN 109
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 35.4 bits (80), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/46 (46%), Positives = 28/46 (61%), Gaps = 2/46 (4%)
Query 1 LFLDRLRKDYSKQIRHWFVC-EFGTLYGRPHYHGILFDVPQTLIDG 45
LFL RLR S +F C E+GT RPHYH ++F+ +L+DG
Sbjct 77 LFLKRLRDRISPHKIRYFGCGEYGTKLQRPHYHLLIFNY-DSLLDG 121
> pss:102457184 XRCC3; X-ray repair complementing defective repair
in Chinese hamster cells 3
Length=349
Score = 37.0 bits (84), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 38/150 (25%), Positives = 66/150 (44%), Gaps = 30/150 (20%)
Query 40 QTLID---GYSPDVPGHHPLLASRWKYG-FVFVGYVSEETCSY--ITKYVTKSINGDKVR 93
Q LID + DVP + + K+G +F+ +V++ Y ITK +T ++ VR
Sbjct 148 QQLIDQQHKFRADVPLD---VIQKIKFGNSIFIEHVADLDTFYNCITKRITLLLSRGMVR 204
Query 94 PRIISSFG-------------IGSNYLDTEEAVLHKLGNQHYQPFMVLNGFQQAMPRYYY 140
+I S + YL T A LH L +++ P + +N A+
Sbjct 205 LVVIDSMAALFRGEFGAKDSVTKAKYLQTFGAKLHSLSSRYRTPILCINQVTDAVDE--- 261
Query 141 NKIFSDVDKQNI-VLDRFINPPVEFSWQGQ 169
+DV + N +LD+ ++P + +W Q
Sbjct 262 ----TDVGQSNFGLLDKNVSPALGLTWSNQ 287
> eclo:ENC_21200 hypothetical protein
Length=378
Score = 36.6 bits (83), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 42/178 (24%), Positives = 65/178 (37%), Gaps = 40/178 (22%)
Query 21 EFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASRWKYGFVFVGYVSEETCSYIT 80
E GR + +F+ +L+ S ++ R Y F + + E
Sbjct 155 EMSKKSGRSNNDKNIFNSITSLMKAQSYELTSLSSRKKERSVYFFHLMSLIDSEM----- 209
Query 81 KYVTKSINGDKVRPRIISSFGIGSNYLDTEEAVLHKLGNQHYQPFMVLNGFQ------QA 134
+ GD++ P S I SNY+ E+ K+ FM LNGF Q
Sbjct 210 --ILLDCEGDEIHPSETESHTIISNYIINGESTASKIN------FMTLNGFLTNEIYYQK 261
Query 135 MPRYY-----------YNKIFSDVDKQNIVLDRFINPPVEFSWQGQKFSSKLERDEMR 181
+ YY YN FS++DK+ I+ W K+ +KL + MR
Sbjct 262 LHEYYVTYINKCFSNFYNDAFSNIDKRKILTKEL--------W--HKYGAKLSLEVMR 309
> lgv:LCGL_0019 hypothetical protein
Length=280
Score = 36.2 bits (82), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 27/82 (33%), Positives = 39/82 (48%), Gaps = 14/82 (17%)
Query 18 FVCEFGTLYGRPHYHGILFDVPQ----TLIDGYSPDVP------GHHPLLASRWKYGFVF 67
FV EF GR H+HG++ P+ TL + +P L W+YGF
Sbjct 118 FVPEFHK-SGRVHFHGVI-GQPEYRSFTLTEATNPKTGEILIRHNRQVLELKEWRYGFTD 175
Query 68 VGYVS--EETCSYITKYVTKSI 87
V + E T SY+TKY++K +
Sbjct 176 VTPIEDKERTSSYMTKYISKEL 197
> lgr:LCGT_0019 hypothetical protein
Length=280
Score = 36.2 bits (82), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 27/82 (33%), Positives = 39/82 (48%), Gaps = 14/82 (17%)
Query 18 FVCEFGTLYGRPHYHGILFDVPQ----TLIDGYSPDVP------GHHPLLASRWKYGFVF 67
FV EF GR H+HG++ P+ TL + +P L W+YGF
Sbjct 118 FVPEFHK-SGRVHFHGVI-GQPEYRSFTLTEATNPKTGEILIRHNRQVLELKEWRYGFTD 175
Query 68 VGYVS--EETCSYITKYVTKSI 87
V + E T SY+TKY++K +
Sbjct 176 VTPIEDKERTSSYMTKYISKEL 197
> mhi:Mhar_0629 adenine deaminase
Length=580
Score = 36.2 bits (82), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 19/58 (33%), Positives = 28/58 (48%), Gaps = 10/58 (17%)
Query 30 HYHGILFDVPQTLI----------DGYSPDVPGHHPLLASRWKYGFVFVGYVSEETCS 77
+Y G++F P L DG+ P + GH PLL+ R +V VG S+ C+
Sbjct 169 NYPGVIFRDPSVLAKICLAGMRRKDGHRPRIDGHSPLLSGRDLAAYVAVGIGSDHECT 226
> lso:CKC_03540 hypothetical protein
Length=107
Score = 34.3 bits (77), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 35/107 (33%), Positives = 47/107 (44%), Gaps = 15/107 (14%)
Query 1 LFLDRLRKDYSKQIRHWFVCEFGTLYGRPHYHGILFDVPQTLIDGYSPDVPGHHPLLASR 60
LFL RLRK+ SK+ R++FV E H G L LI D +
Sbjct 11 LFLKRLRKNTSKKFRYFFVFE-------KHKSGNLH--AHMLIHQEIGDELLKKAEVQEE 61
Query 61 W-KYGFVFVGYVSEE--TCSYITKYVTKSINGDKVRPRIISSFGIGS 104
W + GF V + E+ T Y+ KY+ K D R+ +SF GS
Sbjct 62 WMREGFSHVRLLKEDLNTARYVCKYLLKE---DAKGIRVRASFRYGS 105
> coc:Coch_1957 coproporphyrinogen III oxidase (EC:1.3.99.22)
Length=454
Score = 35.8 bits (81), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 36/79 (46%), Gaps = 0/79 (0%)
Query 144 FSDVDKQNIVLDRFINPPVEFSWQGQKFSSKLERDEMRRSTLNQNITSGLTPALPLPHTE 203
S + K N+ R+ + P W FS++ + +R+S N T G++ + LP E
Sbjct 3 ISLIQKYNVAGPRYTSYPTVPYWDSSTFSTEKWLESLRKSYAESNSTEGISLYIHLPFCE 62
Query 204 RVSSFNRFKKNMDKNKEFK 222
+ +F K + K E +
Sbjct 63 SLCTFCGCHKRITKRHEME 81
Lambda K H a alpha
0.322 0.140 0.435 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 282809995864